library(BEST)
par1 <- as.numeric(par1) #column number of first sample
par2 <- as.numeric(par2) #column number of second sample
par3 <- as.numeric(par3)
par4 <- as.numeric(par4)
if(par6=='yes') {
par7 <- as.numeric(par7)
par8 <- as.numeric(par8)
par9 <- as.numeric(par9)
par10 <- as.numeric(par10)
par11 <- as.numeric(par11)
par12 <- as.numeric(par12)
par13 <- as.numeric(par13)
par14 <- as.numeric(par14)
par15 <- as.numeric(par15)
par16 <- as.numeric(par16)
}
z <- t(y)
if (par1 == par2) stop('Please, select two different column numbers')
if (par1 < 1) stop('Please, select a column number greater than zero for the first sample')
if (par2 < 1) stop('Please, select a column number greater than zero for the second sample')
if (par1 > length(z[1,])) stop('The column number for the first sample should be smaller')
if (par2 > length(z[1,])) stop('The column number for the second sample should be smaller')
if(par6=='no') {
if(par5=='unpaired') {
(r <- BESTmcmc(z[,par1],z[,par2], parallel=F))
}
if(par5=='paired') {
(r <- BESTmcmc(z[,par1]-z[,par2], parallel=F))
}
} else {
yy <- cbind(z[1,],z[2,])
if(par5=='unpaired') {
(r <- BESTmcmc(z[,par1],z[,par2], priors=list(muM = c(par7,par8), muSD = c(par9,par10), sigmaMode = c(par11,par12), sigmaSD = c(par13,par14), nuMean = par15, nuSD = par16), parallel=F))
}
if(par5=='paired') {
(r <- BESTmcmc(z[,par1]-z[,par2], priors=list(muM = c(par7,par8), muSD = c(par9,par10), sigmaMode = c(par11,par12), sigmaSD = c(par13,par14), nuMean = par15, nuSD = par16), parallel=F))
}
}
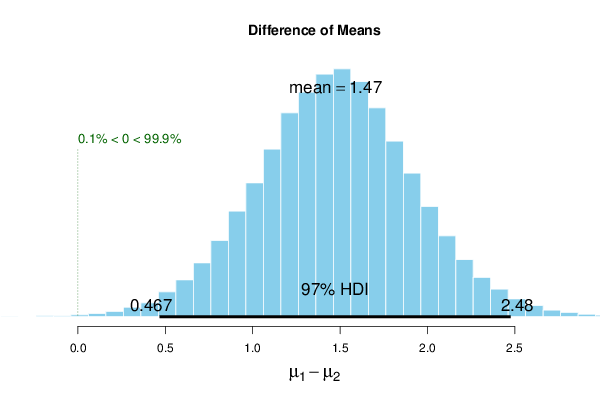
bitmap(file='test2.png')
plot(r, credMass=par4, compVal=par3)
dev.off()
load(file='createtable')
a<-table.start()
a<-table.row.start(a)
a<-table.element(a,'Bayesian Two Sample Test',1,TRUE)
a<-table.row.end(a)
a<-table.row.start(a)
a<-table.element(a,paste('',RC.texteval('summary(r, credMass=par4, compValeff=par3)'),'',sep=''))
a<-table.row.end(a)
a<-table.end(a)
table.save(a,file='mytable.tab')
|