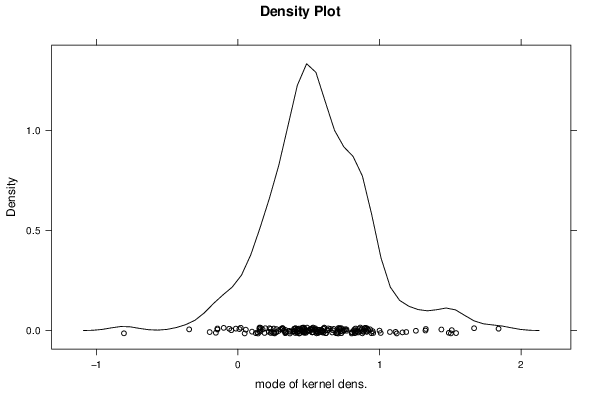
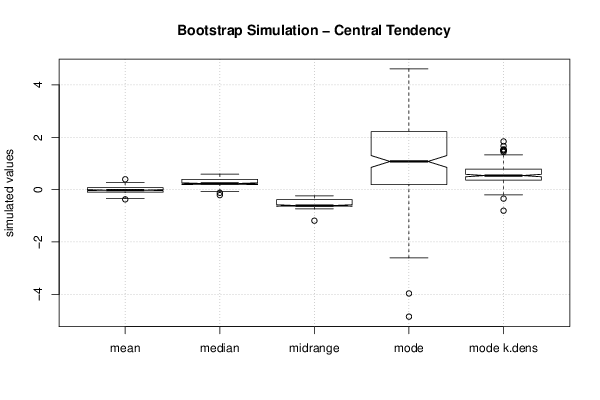
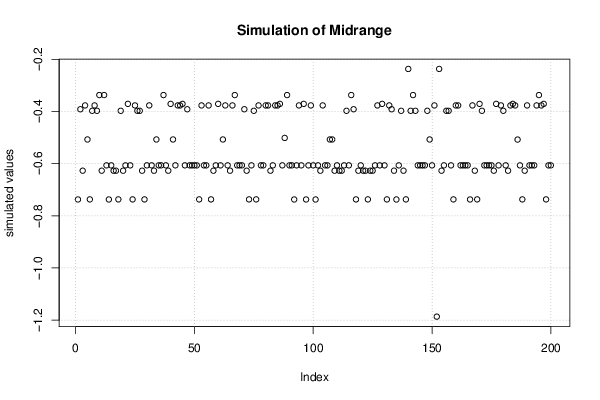
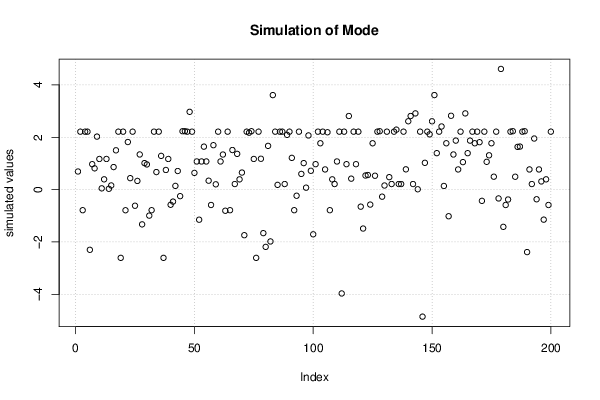
3,23507
0,265899
0,685995
-4,51963
-3,66513
-0,873463
2,71259
0,25908
-0,69719
-3,43973
-1,00121
-4,29834
0,468763
-0,945874
1,30822
-0,867992
-0,0189584
-1,24503
-3,16102
-1,57429
-4,69989
0,609592
-0,913137
1,55224
0,194162
-2,61533
-0,367329
0,623841
-1,1467
-0,0742932
-2,59339
-0,234101
-0,87143
-3,73877
-1,39391
-0,164323
-0,994389
-0,0804383
-3,25867
-1,58861
-5,54757
-0,572814
0,552778
-0,0668002
-0,162364
-1,74025
-0,367992
-5,34741
2,96674
0,185995
-1,48211
-4,54825
-0,494389
2,10185
1,45378
-0,379597
-5,01281
3,83487
0,222917
2,51378
-1,00465
0,0261874
-4,58246
2,35808
-1,00465
-1,40619
-2,45337
0,47207
1,17985
0,640175
-1,47996
0,326705
-0,818791
0,671683
1,19349
0,552778
1,27407
1,29249
-2,84876
-4,2341
-2,00121
1,97902
-1,4213
-3,81238
-3,06201
-0,367318
-0,413007
-2,1152
-3,18079
2,56709
1,69214
-4,85301
-0,327089
-2,1467
-4,60905
-1,2731
-1,95309
0,0323325
2,61224
-5,6152
-0,0134873
-0,486222
-1,63975
-3,05452
1,83764
-2,67262
0,651106
-4,39991
-3,88027
-2,58246
-0,127606
0,539826
-3,68875
1,62864
1,2146
2,11444
1,51364
1,61237
0,214597
2,2146
-0,785403
3,51221
0,573213
4,83225
-2,18667
-0,186672
0,935135
-1,48636
1,07321
1,31333
0,0927902
-0,52694
1,49264
-0,548594
1,93306
-0,428863
3,41348
-2,88829
0,573213
1,01364
0,612367
0,393905
-2,10609
0,97306
0,614443
0,97306
-1,48636
0,413482
-2,60609
0,893905
-5,62709
1,59279
0,893905
1,11444
3,61237
1,71252
1,33168
2,81333
0,913482
0,291675
2,63071
1,67129
0,214597
4,61171
2,07321
2,2321
0,831676
2,31333
0,59279
-0,787479
0,130715
3,01364
0,0142898
1,07652
1,07652
2,51364
-0,586518
2,2146
-0,385557
-1,58859
-0,828709
0,171945
2,2146
-0,307056
-4,92679
1,01364
3,41348
-1,7279
-0,428209
1,11171
2,31125
1,67194
-1,38763
1,23087
2,61237
2,71252
-2,60609
0,812674
2,97098
0,692944
1,89391
2,61237
-0,98571
0,772098
-2,3064
3,61237
-0,488441
4,87348
0,431829
1,51364
3,63194
0,193598
-1,82806
1,53321
0,614443
-1,32806
1,91348
1,47306
2,16987
0,772098
0,393905
-1,33013
0,713175
-1,2854
3,35598
1,82895
0,214597
4,61237
2,37225
0,752521
0,252521
2,2146
2,11237
-2,88556
-0,808478
-4,00944
1,51156
-3,2854
0,0332126
-1,82871
-0,586518
-2,82871
1,17194
2,07321
1,83291
1,81333
0,714597
-2,44844
-0,785403
-2,38556
-3,88556
-0,866634
3,29168
2,07114
-2,06486
2,91348
-1,98779
-3,96679
-0,186672
-6,08652
2,2321
2,51221
-0,566941
1,17194
1,7721
1,47098
3,43306
-1,26913
2,91348
2,2321
3,71317
0,533213
1,81267
2,19294
0,530561
1,81333
1,7721
2,41471 |