Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_smp.wasp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
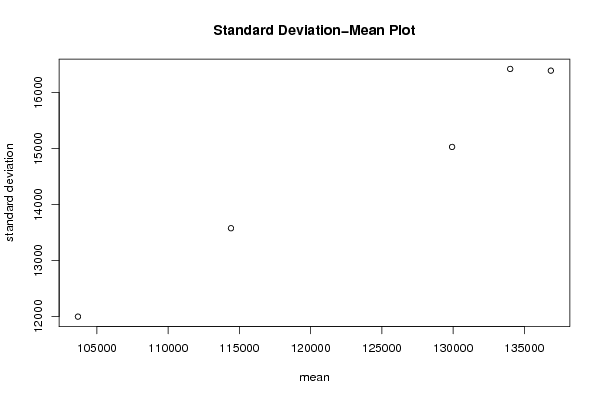
| Title produced by software | Standard Deviation-Mean Plot | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Mon, 08 Dec 2008 05:41:29 -0700 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2008/Dec/08/t12287401620xngpmrof1aytbm.htm/, Retrieved Thu, 16 May 2024 17:44:16 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=30437, Retrieved Thu, 16 May 2024 17:44:16 +0000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | SMP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 209 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| - [Univariate Data Series] [data set] [2008-12-01 19:54:57] [b98453cac15ba1066b407e146608df68] F RMP [Standard Deviation-Mean Plot] [q1] [2008-12-08 12:37:39] [3ffd109c9e040b1ae7e5dbe576d4698c] F D [Standard Deviation-Mean Plot] [SMP] [2008-12-08 12:41:29] [962e6c9020896982bc8283b8971710a9] [Current] - RMP [(Partial) Autocorrelation Function] [ACF] [2008-12-08 12:44:57] [3ffd109c9e040b1ae7e5dbe576d4698c] - RM [Variance Reduction Matrix] [VRM] [2008-12-08 13:10:17] [3ffd109c9e040b1ae7e5dbe576d4698c] - RM [(Partial) Autocorrelation Function] [ACF] [2008-12-08 13:14:12] [3ffd109c9e040b1ae7e5dbe576d4698c] F [(Partial) Autocorrelation Function] [ACF] [2008-12-08 13:16:18] [3ffd109c9e040b1ae7e5dbe576d4698c] - P [(Partial) Autocorrelation Function] [ACF met meer lags] [2008-12-15 14:07:02] [f77c9ab3b413812d7baee6b7ec69a15d] - P [(Partial) Autocorrelation Function] [ACF] [2008-12-16 11:53:49] [3ffd109c9e040b1ae7e5dbe576d4698c] - R P [(Partial) Autocorrelation Function] [ACF] [2008-12-18 16:25:12] [3ffd109c9e040b1ae7e5dbe576d4698c] - P [(Partial) Autocorrelation Function] [ACF] [2008-12-24 12:57:31] [b28ef2aea2cd58ceb5ad90223572c703] F [(Partial) Autocorrelation Function] [ACF] [2008-12-08 13:17:45] [3ffd109c9e040b1ae7e5dbe576d4698c] F RM [Spectral Analysis] [spectraal] [2008-12-08 13:20:34] [3ffd109c9e040b1ae7e5dbe576d4698c] - R P [Spectral Analysis] [Spectraal] [2008-12-18 16:27:12] [3ffd109c9e040b1ae7e5dbe576d4698c] - [Spectral Analysis] [spectraal analyse] [2008-12-24 12:59:19] [b28ef2aea2cd58ceb5ad90223572c703] - R P [Spectral Analysis] [spectraal] [2008-12-18 18:22:50] [3ffd109c9e040b1ae7e5dbe576d4698c] - RM [Spectral Analysis] [spectraal] [2008-12-08 13:22:13] [3ffd109c9e040b1ae7e5dbe576d4698c] - R P [Spectral Analysis] [spectraal] [2008-12-18 18:23:46] [3ffd109c9e040b1ae7e5dbe576d4698c] - [Spectral Analysis] [spectraal analyse] [2008-12-24 13:02:50] [b28ef2aea2cd58ceb5ad90223572c703] - P [Spectral Analysis] [spectraal] [2008-12-24 13:21:21] [3ffd109c9e040b1ae7e5dbe576d4698c] - RM [Spectral Analysis] [spectraal] [2008-12-08 13:23:27] [3ffd109c9e040b1ae7e5dbe576d4698c] F RM [(Partial) Autocorrelation Function] [ACF] [2008-12-08 13:40:41] [3ffd109c9e040b1ae7e5dbe576d4698c] - R P [(Partial) Autocorrelation Function] [acf] [2008-12-18 18:26:49] [3ffd109c9e040b1ae7e5dbe576d4698c] - P [(Partial) Autocorrelation Function] [autocorrelatie en...] [2008-12-19 10:44:18] [3f66c6f083b1153972739491b89fa2dd] - [Spectral Analysis] [spectraal] [2008-12-08 13:42:55] [3ffd109c9e040b1ae7e5dbe576d4698c] - R P [Spectral Analysis] [spectraal] [2008-12-18 18:28:37] [3ffd109c9e040b1ae7e5dbe576d4698c] - R P [Spectral Analysis] [spectraal] [2008-12-18 18:25:11] [3ffd109c9e040b1ae7e5dbe576d4698c] - P [Spectral Analysis] [spectraal analyse] [2008-12-24 13:07:13] [b28ef2aea2cd58ceb5ad90223572c703] - P [(Partial) Autocorrelation Function] [ACF met meer lags] [2008-12-15 14:18:46] [f77c9ab3b413812d7baee6b7ec69a15d] - R P [(Partial) Autocorrelation Function] [ACF] [2008-12-18 16:54:54] [3ffd109c9e040b1ae7e5dbe576d4698c] - P [(Partial) Autocorrelation Function] [ACF] [2008-12-24 13:04:28] [b28ef2aea2cd58ceb5ad90223572c703] - R P [(Partial) Autocorrelation Function] [ACF] [2008-12-18 16:48:38] [3ffd109c9e040b1ae7e5dbe576d4698c] - P [(Partial) Autocorrelation Function] [ACF] [2008-12-24 13:00:55] [b28ef2aea2cd58ceb5ad90223572c703] - R [Variance Reduction Matrix] [VRM] [2008-12-18 18:18:02] [3ffd109c9e040b1ae7e5dbe576d4698c] - R [Standard Deviation-Mean Plot] [plot] [2008-12-18 18:16:11] [3ffd109c9e040b1ae7e5dbe576d4698c] - [Standard Deviation-Mean Plot] [plot] [2008-12-24 12:54:32] [b28ef2aea2cd58ceb5ad90223572c703] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
147768 137507 136919 136151 133001 125554 119647 114158 116193 152803 161761 160942 149470 139208 134588 130322 126611 122401 117352 112135 112879 148729 157230 157221 146681 136524 132111 125326 122716 116615 113719 110737 112093 143565 149946 149147 134339 122683 115614 116566 111272 104609 101802 94542 93051 124129 130374 123946 114971 105531 104919 104782 101281 94545 93248 84031 87486 115867 120327 117008 108811 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
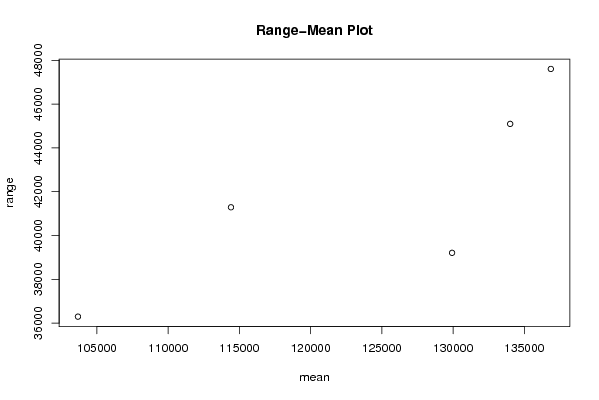
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 12 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 12 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
par1 <- as.numeric(par1) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||