Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||
| R Software Module | rwasp_meanversusmedian.wasp | ||||||||||||||||||||||||||||||||
| Title produced by software | Mean versus Median | ||||||||||||||||||||||||||||||||
| Date of computation | Wed, 21 Dec 2011 12:18:05 -0500 | ||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2011/Dec/21/t13244881241bch4380zb0hj40.htm/, Retrieved Wed, 17 Sep 2025 02:05:01 +0000 | ||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=158899, Retrieved Wed, 17 Sep 2025 02:05:01 +0000 | |||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||
| Estimated Impact | 199 | ||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||
| - [Univariate Explorative Data Analysis] [Run Sequence gebo...] [2008-12-12 13:32:37] [76963dc1903f0f612b6153510a3818cf] - R D [Univariate Explorative Data Analysis] [Run Sequence gebo...] [2008-12-17 12:14:40] [76963dc1903f0f612b6153510a3818cf] - [Univariate Explorative Data Analysis] [Run Sequence Plot...] [2008-12-22 18:19:51] [1ce0d16c8f4225c977b42c8fa93bc163] - RMP [Univariate Data Series] [Identifying Integ...] [2009-11-22 12:08:06] [b98453cac15ba1066b407e146608df68] - RMP [Classical Decomposition] [ws8. additional] [2011-11-30 13:03:24] [8ae0a4da1b3ee81f40dbba5e42914d07] - RMP [Exponential Smoothing] [WS8 Monthly birth...] [2011-11-30 14:31:27] [2628f630b839c9d14ba9c3627ab0414a] - RMPD [Mean versus Median] [Paper] [2011-12-21 17:18:05] [3d1c016f2f3d502d2cccf121bc4326eb] [Current] | |||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||
-31.0980235042716 -59.8206924271108 -83.5404637229203 -76.6837390525943 -55.7725703639553 -29.62380040276 -403.243804757836 -416.782395531722 -213.421552958589 371.566721695855 165.983275906532 542.6202623301 295.02721709437 -21.1190278168924 437.4008577275 365.294800512585 67.9306326630958 -247.788856282479 545.098074661517 -296.773620935714 352.477617436036 283.798200110577 -273.315875572704 446.06222457008 -214.72781956204 -60.4206198854663 372.155927347334 99.2591381678085 318.424339918272 125.277834109822 -327.853683550757 -432.970794551573 442.139159285423 14.3549856459813 299.040533753827 88.2365194871691 -180.537099245963 201.985172786433 193.536881410406 -313.092895033498 -152.748572628851 299.982624154345 42.4811421719933 133.842278481663 -129.830065090609 -699.373730899397 171.563382431515 -24.5384434740972 116.363164839684 -218.154308468131 -257.102541429251 98.6124781717281 -190.742241794984 198.422707475118 -145.26733998686 611.517071444689 56.0522598394018 -117.514342950461 -3.71499521678197 296.99038945692 360.610007278467 61.6319235909414 -746.579530943922 | |||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||
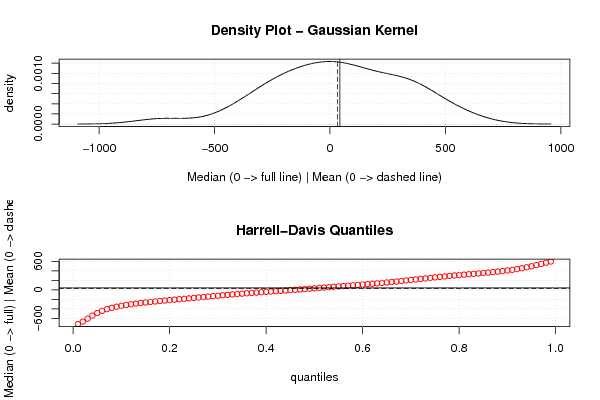
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||
| par1 = 12 ; | |||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||
library(Hmisc) | |||||||||||||||||||||||||||||||||