Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||
| R Software Module | rwasp_meanversusmedian.wasp | ||||||||||||||||||||||||||||||||
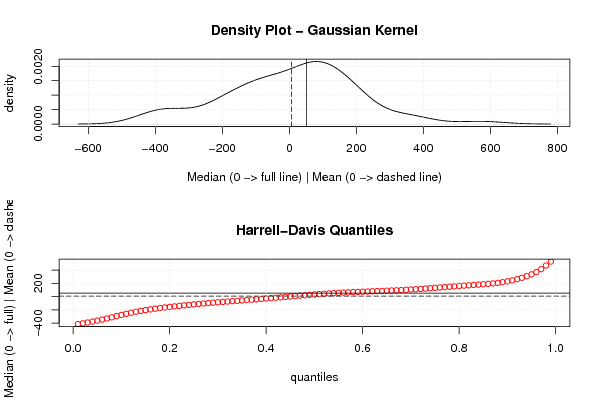
| Title produced by software | Mean versus Median | ||||||||||||||||||||||||||||||||
| Date of computation | Wed, 21 Dec 2011 09:15:06 -0500 | ||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2011/Dec/21/t13244771092mywut796ym5r1o.htm/, Retrieved Sat, 25 Oct 2025 16:54:22 +0000 | ||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=158720, Retrieved Sat, 25 Oct 2025 16:54:22 +0000 | |||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||
| Estimated Impact | 276 | ||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||
| - [Univariate Explorative Data Analysis] [Run Sequence gebo...] [2008-12-12 13:32:37] [76963dc1903f0f612b6153510a3818cf] - R D [Univariate Explorative Data Analysis] [Run Sequence gebo...] [2008-12-17 12:14:40] [76963dc1903f0f612b6153510a3818cf] - [Univariate Explorative Data Analysis] [Run Sequence Plot...] [2008-12-22 18:19:51] [1ce0d16c8f4225c977b42c8fa93bc163] - RMP [Univariate Data Series] [Identifying Integ...] [2009-11-22 12:08:06] [b98453cac15ba1066b407e146608df68] - RMP [Classical Decomposition] [ws8. additional] [2011-11-30 13:03:24] [8ae0a4da1b3ee81f40dbba5e42914d07] - [Classical Decomposition] [WS8 Monthly births] [2011-11-30 13:18:52] [2628f630b839c9d14ba9c3627ab0414a] - RMPD [Mean versus Median] [Paper blog] [2011-12-21 14:15:06] [3d1c016f2f3d502d2cccf121bc4326eb] [Current] | |||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||
251.97013888889 572.861805555554 -37.4798611111109 51.2284722222212 23.2534722222226 -382.704861111111 97.478472222223 168.111805555556 -207.379861111111 9.66180555555547 68.6118055555562 90.6784722222237 -151.90486111111 91.5701388888901 -398.563194444443 191.811805555557 -38.0798611111095 -62.8715277777774 185.936805555557 -77.013194444442 -43.6715277777766 115.245138888889 -75.888194444442 -358.113194444444 354.845138888888 -103.429861111112 -15.8548611111109 336.145138888889 -338.454861111111 103.003472222223 -167.104861111109 -126.67986111111 138.703472222223 80.4534722222234 208.653472222222 -89.2381944444423 -138.696527777776 -422.80486111111 207.395138888887 193.478472222223 70.7118055555566 66.2951388888905 -217.938194444443 88.4034722222241 177.411805555555 -224.796527777777 -47.138194444442 157.178472222224 80.095138888888 6.82013888889014 92.0618055555569 -339.104861111111 166.711805555557 55.1701388888887 51.3118055555569 -103.138194444444 -115.379861111112 -30.8798611111106 -204.55486111111 149.178472222224 -194.65486111111 377.528472222222 64.6451388888909 | |||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||
library(Hmisc) | |||||||||||||||||||||||||||||||||