Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_smp.wasp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
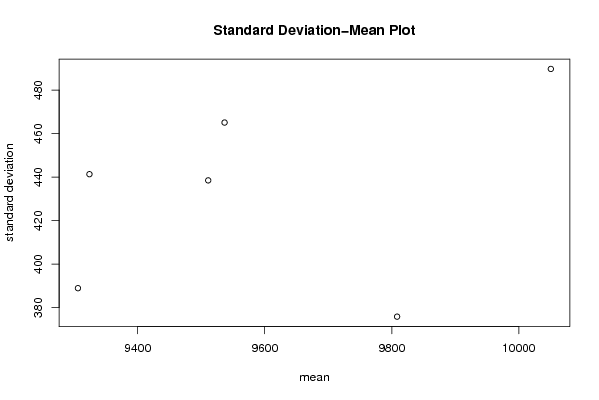
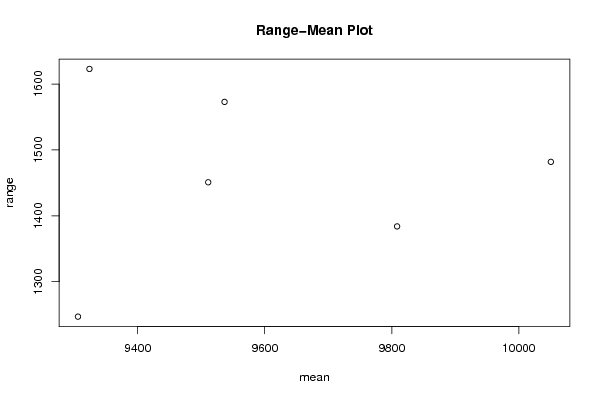
| Title produced by software | Standard Deviation-Mean Plot | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Mon, 29 Nov 2010 10:52:49 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2010/Nov/29/t1291027857lvys8qncifu969u.htm/, Retrieved Wed, 17 Sep 2025 14:13:00 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=102825, Retrieved Wed, 17 Sep 2025 14:13:00 +0000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 800 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| - [Univariate Explorative Data Analysis] [Run Sequence gebo...] [2008-12-12 13:32:37] [76963dc1903f0f612b6153510a3818cf] - R D [Univariate Explorative Data Analysis] [Run Sequence gebo...] [2008-12-17 12:14:40] [76963dc1903f0f612b6153510a3818cf] - [Univariate Explorative Data Analysis] [Run Sequence Plot...] [2008-12-22 18:19:51] [1ce0d16c8f4225c977b42c8fa93bc163] - RMP [Univariate Data Series] [Identifying Integ...] [2009-11-22 12:08:06] [b98453cac15ba1066b407e146608df68] - RMP [Standard Deviation-Mean Plot] [Births] [2010-11-29 10:52:49] [d76b387543b13b5e3afd8ff9e5fdc89f] [Current] - RMP [ARIMA Backward Selection] [Births] [2010-11-29 17:42:52] [b98453cac15ba1066b407e146608df68] F R PD [ARIMA Backward Selection] [ARIMA] [2010-12-02 20:40:17] [97ad38b1c3b35a5feca8b85f7bc7b3ff] - R P [ARIMA Backward Selection] [] [2011-12-04 15:27:05] [9401a40688cf36283be626153bc5a38b] - R [ARIMA Backward Selection] [WS9.7] [2011-12-06 20:38:53] [246430c28552f7bc561843533d7ec653] - RMP [ARIMA Backward Selection] [WS 9 - ARIMA para...] [2011-12-05 14:17:13] [74be16979710d4c4e7c6647856088456] - R PD [ARIMA Backward Selection] [] [2011-12-05 18:48:35] [06c08141d7d783218a8164fd2ea166f2] - R PD [ARIMA Backward Selection] [] [2011-12-20 19:48:55] [06c08141d7d783218a8164fd2ea166f2] - R PD [ARIMA Backward Selection] [C9.7] [2011-12-05 19:02:36] [d1ce18d003fa52f731d1c3ce8b58d5f9] - R P [ARIMA Backward Selection] [c9.7] [2011-12-05 19:22:18] [d1ce18d003fa52f731d1c3ce8b58d5f9] - PD [ARIMA Backward Selection] [Model 1: CPI] [2010-12-04 12:17:27] [6bc4f9343b7ea3ef5a59412d1f72bb2b] - PD [ARIMA Backward Selection] [Model 2: CPI] [2010-12-04 12:20:20] [6bc4f9343b7ea3ef5a59412d1f72bb2b] - PD [ARIMA Backward Selection] [Test 5] [2010-12-05 11:00:47] [74be16979710d4c4e7c6647856088456] - P [ARIMA Backward Selection] [W9 - Blog 8] [2010-12-06 16:09:21] [1aa8d85d6b335d32b1f6be940e33a166] - D [ARIMA Backward Selection] [Blog 4] [2010-12-10 11:39:46] [1aa8d85d6b335d32b1f6be940e33a166] - PD [ARIMA Backward Selection] [ARMA parameters] [2010-12-21 13:20:08] [74be16979710d4c4e7c6647856088456] - [ARIMA Backward Selection] [Arima backward] [2010-12-21 19:45:42] [717f3d787904f94c39256c5c1fc72d4c] - PD [ARIMA Backward Selection] [ARMA parameters] [2010-12-21 20:32:57] [74be16979710d4c4e7c6647856088456] - [ARIMA Backward Selection] [Arima backward] [2010-12-21 20:47:35] [717f3d787904f94c39256c5c1fc72d4c] - [ARIMA Backward Selection] [ws 9] [2011-12-07 11:14:31] [27e29806e0b1d1351a97bc4ee4116294] F P [ARIMA Backward Selection] [W9 3] [2010-12-06 18:22:48] [1afa3497b02a8d7c9f6727c1b17b89b2] F PD [ARIMA Backward Selection] [W9] [2010-12-06 19:00:10] [5ddc7dfb25e070b079c4c8fcccc4d42e] - [ARIMA Backward Selection] [] [2010-12-15 15:13:49] [2e1e44f0ae3cb9513dc28781dfdb387b] - [ARIMA Backward Selection] [] [2010-12-17 16:50:02] [dc73d270d5d96f29ff77294e1b86f79b] - P [ARIMA Backward Selection] [ARMA parameters] [2011-12-07 12:53:48] [489eb911c8db04aca1fc54d886fc3144] - P [ARIMA Backward Selection] [ws9.6] [2011-12-07 13:25:00] [8ae0a4da1b3ee81f40dbba5e42914d07] - [ARIMA Backward Selection] [ARMA parameter] [2011-12-08 08:41:51] [d1852ae60c447c0faaa37b55ffb3cb2b] - RMP [ARIMA Forecasting] [Forecasting] [2011-12-08 08:48:03] [d1852ae60c447c0faaa37b55ffb3cb2b] - [ARIMA Backward Selection] [ARIMA parameters ...] [2011-12-08 14:21:22] [d5821fed422662f85834b1edae505ce2] - [ARIMA Backward Selection] [workshop 9g] [2011-12-08 14:32:47] [bab4e1d4a779bb46523d87231e2a2e96] - D [ARIMA Backward Selection] [workshop 9] [2011-12-08 14:34:12] [b1d8f9e536fbb1cbb331f583f7f5f24b] - [ARIMA Backward Selection] [ARIMA backward] [2011-12-22 13:41:46] [bab4e1d4a779bb46523d87231e2a2e96] - R [ARIMA Backward Selection] [] [2012-11-26 21:01:37] [74be16979710d4c4e7c6647856088456] F PD [ARIMA Backward Selection] [ARIMA] [2010-12-06 20:02:38] [2843717cd92615903379c14ebee3c5df] - PD [ARIMA Backward Selection] [WS 9 ] [2010-12-06 21:01:55] [19f9551d4d95750ef21e9f3cf8fe2131] F PD [ARIMA Backward Selection] [ws 9] [2010-12-07 10:21:33] [bd591a1ebb67d263a02e7adae3fa1a4d] - RMPD [ARIMA Forecasting] [] [2010-12-14 12:31:06] [9b13650c94c5192ca5135ec8a1fa39f7] - [ARIMA Backward Selection] [arima] [2010-12-21 10:06:40] [bd591a1ebb67d263a02e7adae3fa1a4d] - PD [ARIMA Backward Selection] [arima 2] [2010-12-22 18:11:01] [bd591a1ebb67d263a02e7adae3fa1a4d] - [ARIMA Backward Selection] [] [2011-12-04 14:29:55] [50e3859e0b739a5118d466e989dfc0cb] - R P [ARIMA Backward Selection] [] [2011-12-04 14:31:26] [50e3859e0b739a5118d466e989dfc0cb] - RMPD [Skewness and Kurtosis Test] [] [2011-12-04 14:59:57] [50e3859e0b739a5118d466e989dfc0cb] - R [ARIMA Backward Selection] [ARIMA- Births] [2011-12-07 13:16:21] [74be16979710d4c4e7c6647856088456] - PD [ARIMA Backward Selection] [paper ARIMA (fout...] [2011-12-09 13:07:59] [7e261c986c934df955dd3ac53e9d45c6] - R P [ARIMA Backward Selection] [paper - ARIMA] [2011-12-11 10:44:54] [7e261c986c934df955dd3ac53e9d45c6] - R PD [ARIMA Backward Selection] [ARIMA backward se...] [2011-12-18 12:40:22] [379dab8110dbf77cfcc4b7951c3a599f] - P [ARIMA Backward Selection] [ARIMA model] [2012-11-22 09:40:31] [0dc867bfbaab36a894719867823e3cb9] - R P [ARIMA Backward Selection] [WS9-7] [2012-11-29 19:50:13] [f4f48270c576c45d216b84daa061a85b] - RMP [ARIMA Forecasting] [WS9-8] [2012-11-29 20:08:15] [f4f48270c576c45d216b84daa061a85b] [Truncated] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
9700 9081 9084 9743 8587 9731 9563 9998 9437 10038 9918 9252 9737 9035 9133 9487 8700 9627 8947 9283 8829 9947 9628 9318 9605 8640 9214 9567 8547 9185 9470 9123 9278 10170 9434 9655 9429 8739 9552 9687 9019 9672 9206 9069 9788 10312 10105 9863 9656 9295 9946 9701 9049 10190 9706 9765 9893 9994 10433 10073 10112 9266 9820 10097 9115 10411 9678 10408 10153 10368 10581 10597 10680 9738 9556 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 1 ; par2 = 0 ; par3 = 0 ; par4 = 12 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 12 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
par1 <- as.numeric(par1) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||