x <- as.data.frame(read.table(file='https://automated.biganalytics.eu/download/statdb.csv',sep=',',header=T))
library(GenKern)
load(file='createtable')
doplot1 <- function(myxcol,myycol,myxlab='',myylab='') {
p0 <- x[x$Pop==0,]
p1 <- x[x$Pop==1,]
dum <- cbind(p0[,myxcol],rank(p0[,myxcol]),p0[,'Gender'])
xf0 <- dum[dum[,3]==0,2]
xm0 <- dum[dum[,3]==1,2]
dum <- cbind(p0[,myycol],rank(p0[,myycol]),p0[,'Gender'])
yf0 <- dum[dum[,3]==0,2]
ym0 <- dum[dum[,3]==1,2]
dum <- cbind(p1[,myxcol],rank(p1[,myxcol]),p1[,'Gender'])
xf1 <- dum[dum[,3]==0,2]
xm1 <- dum[dum[,3]==1,2]
dum <- cbind(p1[,myycol],rank(p1[,myycol]),p1[,'Gender'])
yf1 <- dum[dum[,3]==0,2]
ym1 <- dum[dum[,3]==1,2]
xf <- rank(x[x$Gender==0,myxcol])
yf <- rank(x[x$Gender==0,myycol])
xm <- rank(x[x$Gender==1,myxcol])
ym <- rank(x[x$Gender==1,myycol])
x0 <- rank(x[x$Pop==0,myxcol])
y0 <- rank(x[x$Pop==0,myycol])
x1 <- rank(x[x$Pop==1,myxcol])
y1 <- rank(x[x$Pop==1,myycol])
mycorr <- array(NA,dim=c(4,4))
rownames(mycorr) <- c('Female Bachelor Students','Male Bachelor Students','Female Switching Students','Male Switching Students')
colnames(mycorr) <- c('Pearson rho','Pearson p-value','Kendall tau','Kendall p-value')
bitmap(file='mypicture.png')
par(mfrow=c(2,2))
r <- cor.test(xf0,yf0,method='pearson')
mycorr[1,1] <- r$estimate
mycorr[1,2] <- r$p.value
r <- cor.test(xf0,yf0,method='kendall')
mycorr[1,3] <- r$estimate
mycorr[1,4] <- r$p.value
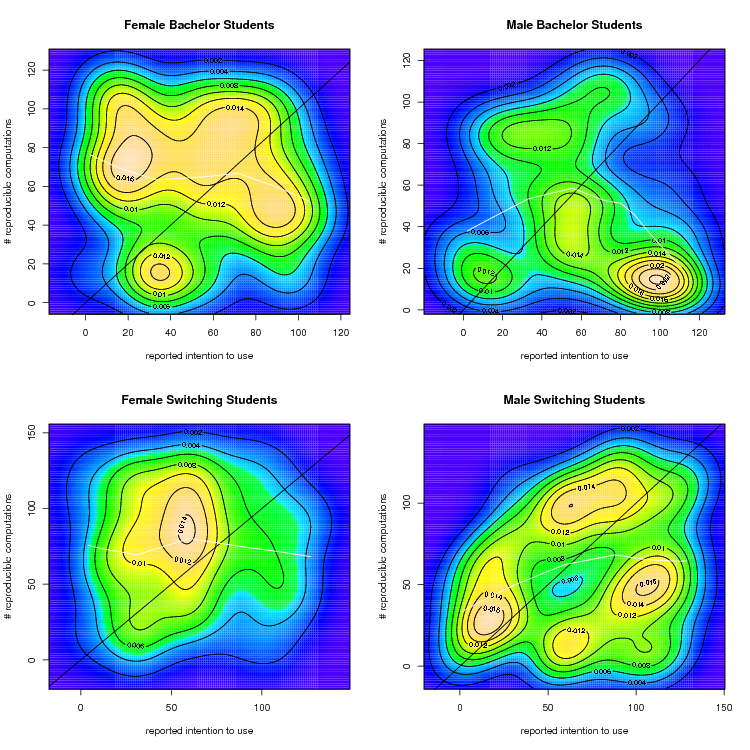
op <- KernSur(xf0,yf0, xgridsize=150, ygridsize=150,na.rm=T)
image(op$xords, op$yords, op$zden, col=topo.colors(100), axes=TRUE, xlab=myxlab, ylab=myylab, main=rownames(mycorr)[1])
contour(op$xords, op$yords, op$zden, add=TRUE)
box()
abline(0,1)
lines(stats::lowess(cbind(xf0,yf0)),col='white')
r <- cor.test(xm0,ym0,method='pearson')
mycorr[2,1] <- r$estimate
mycorr[2,2] <- r$p.value
r <- cor.test(xm0,ym0,method='kendall')
mycorr[2,3] <- r$estimate
mycorr[2,4] <- r$p.value
op <- KernSur(xm0,ym0, xgridsize=150, ygridsize=150,na.rm=T)
image(op$xords, op$yords, op$zden, col=topo.colors(100), axes=TRUE, xlab=myxlab, ylab=myylab, main=rownames(mycorr)[2])
contour(op$xords, op$yords, op$zden, add=TRUE)
box()
abline(0,1)
lines(stats::lowess(cbind(xm0,ym0)),col='white')
r <- cor.test(xf1,yf1,method='pearson')
mycorr[3,1] <- r$estimate
mycorr[3,2] <- r$p.value
r <- cor.test(xf1,yf1,method='kendall')
mycorr[3,3] <- r$estimate
mycorr[3,4] <- r$p.value
op <- KernSur(xf1,yf1, xgridsize=150, ygridsize=150,na.rm=T)
image(op$xords, op$yords, op$zden, col=topo.colors(100), axes=TRUE, xlab=myxlab, ylab=myylab, main=rownames(mycorr)[3])
contour(op$xords, op$yords, op$zden, add=TRUE)
box()
abline(0,1)
lines(stats::lowess(cbind(xf1,yf1)),col='white')
r <- cor.test(xm1,ym1,method='pearson')
mycorr[4,1] <- r$estimate
mycorr[4,2] <- r$p.value
r <- cor.test(xm1,ym1,method='kendall')
mycorr[4,3] <- r$estimate
mycorr[4,4] <- r$p.value
op <- KernSur(xm1,ym1, xgridsize=150, ygridsize=150,na.rm=T)
image(op$xords, op$yords, op$zden, col=topo.colors(100), axes=TRUE, xlab=myxlab, ylab=myylab, main=rownames(mycorr)[4])
contour(op$xords, op$yords, op$zden, add=TRUE)
box()
abline(0,1)
lines(stats::lowess(cbind(xm1,ym1)),col='white')
dev.off()
mycorr
}
if (par1 == 'actual feedback') {
myxcol='nnzfg'
myxlab = '# submitted messages'
}
if (par1 == 'reported feedback') {
myxcol = 'Reflection'
myxlab = 'reported feedback submissions'
}
if (par1 == 'reported computing') {
myxcol = 'Future'
myxlab = 'reported intention to use'
}
if (par2 == 'actual computing') {
myycol = 'Bcount'
myylab = '# reproducible computations'
}
if (par2 == 'actual feedback') {
myycol = 'nnzfg'
myylab = 'actual feedback submissions'
}
if (par2 == 'reported computing') {
myycol = 'Future'
myylab = 'reported intention to use'
}
mycorr <- doplot1(myxcol=myxcol,myycol=myycol,myxlab=myxlab,myylab=myylab)
a<-table.start()
a<-table.row.start(a)
dum <- 'Correlations between'
dum <- paste(dum,myxlab,sep=' ')
dum <- paste(dum,myylab,sep=' and ')
a<-table.element(a,dum,5,TRUE)
a<-table.row.end(a)
a<-table.row.start(a)
a<-table.element(a,'',header=TRUE)
a<-table.element(a,'Pearson rho',header=TRUE)
a<-table.element(a,'Pearson p-value',header=TRUE)
a<-table.element(a,'Kendall tau',header=TRUE)
a<-table.element(a,'Kendall p-value',header=TRUE)
a<-table.row.end(a)
for (myrow in 1:4) {
a<-table.row.start(a)
a<-table.element(a,rownames(mycorr)[myrow],header=TRUE)
a<-table.element(a,round(mycorr[myrow,1],4))
a<-table.element(a,round(mycorr[myrow,2],4))
a<-table.element(a,round(mycorr[myrow,3],4))
a<-table.element(a,round(mycorr[myrow,4],4))
a<-table.row.end(a)
}
a<-table.end(a)
table.save(a,file='mytable.tab')
|