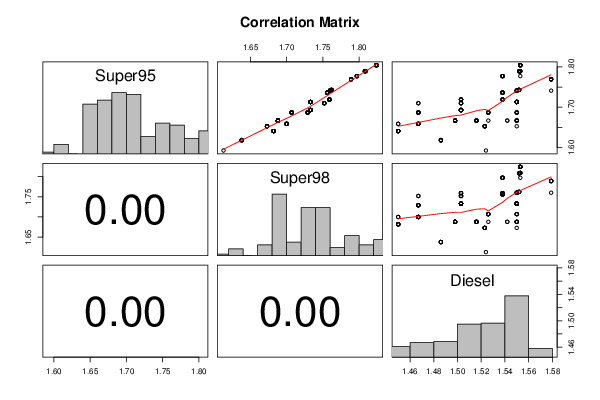
1,593 1,613 1,524
1,619 1,638 1,486
1,619 1,638 1,486
1,618 1,638 1,486
1,618 1,638 1,486
1,618 1,638 1,486
1,653 1,673 1,523
1,653 1,673 1,523
1,653 1,673 1,523
1,653 1,673 1,523
1,653 1,673 1,523
1,653 1,673 1,523
1,653 1,673 1,523
1,653 1,673 1,55
1,667 1,688 1,55
1,667 1,688 1,55
1,667 1,688 1,55
1,667 1,688 1,55
1,667 1,688 1,55
1,667 1,688 1,55
1,667 1,688 1,516
1,667 1,688 1,516
1,667 1,688 1,516
1,667 1,688 1,516
1,667 1,688 1,516
1,667 1,688 1,516
1,667 1,688 1,516
1,667 1,688 1,516
1,667 1,688 1,498
1,667 1,688 1,498
1,667 1,688 1,498
1,667 1,688 1,498
1,667 1,688 1,498
1,667 1,688 1,498
1,667 1,688 1,498
1,667 1,688 1,498
1,667 1,688 1,498
1,667 1,688 1,498
1,667 1,688 1,498
1,667 1,688 1,542
1,667 1,688 1,542
1,667 1,688 1,526
1,687 1,707 1,526
1,687 1,707 1,526
1,687 1,707 1,526
1,687 1,707 1,526
1,687 1,707 1,526
1,687 1,707 1,526
1,687 1,707 1,55
1,687 1,707 1,55
1,687 1,707 1,55
1,687 1,707 1,55
1,713 1,733 1,55
1,713 1,733 1,55
1,713 1,733 1,55
1,713 1,733 1,55
1,713 1,733 1,55
1,713 1,733 1,55
1,713 1,733 1,55
1,713 1,733 1,55
1,713 1,733 1,55
1,713 1,733 1,55
1,713 1,733 1,55
1,713 1,733 1,55
1,713 1,733 1,55
1,713 1,733 1,55
1,741 1,76 1,55
1,741 1,76 1,55
1,741 1,76 1,55
1,741 1,76 1,55
1,741 1,76 1,55
1,741 1,76 1,55
1,741 1,76 1,55
1,741 1,76 1,55
1,741 1,76 1,55
1,741 1,76 1,579
1,769 1,789 1,579
1,769 1,789 1,579
1,769 1,789 1,579
1,769 1,789 1,579
1,769 1,789 1,579
1,769 1,789 1,579
1,769 1,789 1,579
1,769 1,789 1,579
1,743 1,762 1,552
1,743 1,762 1,552
1,743 1,762 1,552
1,743 1,762 1,552
1,743 1,762 1,552
1,743 1,762 1,552
1,789 1,808 1,552
1,789 1,808 1,552
1,789 1,808 1,552
1,789 1,808 1,552
1,789 1,809 1,553
1,789 1,809 1,553
1,789 1,809 1,553
1,789 1,809 1,553
1,804 1,824 1,553
1,804 1,824 1,553
1,804 1,824 1,553
1,804 1,824 1,553
1,804 1,824 1,553
1,804 1,824 1,553
1,804 1,824 1,553
1,804 1,824 1,553
1,804 1,824 1,553
1,804 1,824 1,553
1,804 1,824 1,553
1,804 1,824 1,553
1,777 1,797 1,553
1,777 1,797 1,538
1,777 1,797 1,538
1,777 1,797 1,538
1,777 1,797 1,538
1,777 1,797 1,538
1,777 1,797 1,538
1,736 1,756 1,538
1,736 1,756 1,538
1,736 1,756 1,538
1,736 1,756 1,538
1,736 1,756 1,538
1,736 1,756 1,538
1,736 1,756 1,538
1,736 1,756 1,538
1,736 1,756 1,538
1,719 1,759 1,538
1,719 1,759 1,538
1,719 1,759 1,538
1,719 1,759 1,538
1,719 1,759 1,538
1,719 1,759 1,503
1,719 1,759 1,503
1,719 1,759 1,503
1,719 1,759 1,503
1,719 1,759 1,503
1,693 1,733 1,503
1,693 1,733 1,503
1,693 1,733 1,503
1,693 1,733 1,503
1,693 1,733 1,503
1,693 1,733 1,503
1,693 1,733 1,503
1,693 1,733 1,503
1,693 1,733 1,503
1,693 1,733 1,503
1,693 1,733 1,503
1,693 1,733 1,503
1,693 1,733 1,503
1,693 1,733 1,503
1,693 1,733 1,503
1,693 1,733 1,503
1,71 1,752 1,503
1,71 1,752 1,503
1,71 1,752 1,503
1,71 1,752 1,503
1,71 1,752 1,503
1,71 1,752 1,467
1,71 1,752 1,467
1,687 1,729 1,467
1,687 1,729 1,467
1,687 1,729 1,467
1,687 1,729 1,467
1,687 1,729 1,467
1,687 1,729 1,467
1,659 1,7 1,467
1,659 1,7 1,467
1,659 1,7 1,467
1,659 1,7 1,467
1,659 1,7 1,467
1,659 1,7 1,467
1,659 1,7 1,467
1,659 1,7 1,45
1,659 1,7 1,45
1,641 1,682 1,45
1,641 1,682 1,45
1,641 1,682 1,45
1,641 1,682 1,45
1,641 1,682 1,45
1,641 1,682 1,45
1,641 1,682 1,45
1,641 1,682 1,45
1,641 1,682 1,45
|