Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Author | *Unverified author* | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | -- | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
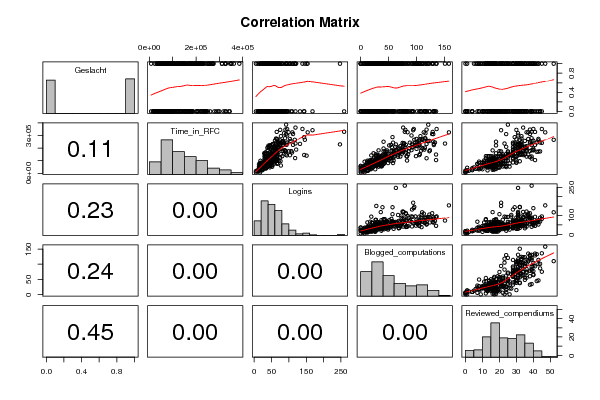
| Title produced by software | Kendall tau Correlation Matrix | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Fri, 23 Dec 2011 14:28:18 -0500 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2011/Dec/23/t1324668693ua9cebo80q6t0g4.htm/, Retrieved Mon, 29 Apr 2024 18:02:53 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=160679, Retrieved Mon, 29 Apr 2024 18:02:53 +0000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 89 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| - [Kendall tau Correlation Matrix] [] [2010-12-05 17:44:33] [b98453cac15ba1066b407e146608df68] - RMPD [Kendall tau Correlation Matrix] [] [2011-12-15 18:41:33] [145a13cc95845961a3828fae7139a7eb] - PD [Kendall tau Correlation Matrix] [] [2011-12-21 13:34:32] [74be16979710d4c4e7c6647856088456] - M [Kendall tau Correlation Matrix] [] [2011-12-23 19:28:18] [d41d8cd98f00b204e9800998ecf8427e] [Current] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
1 210907 56 79 30 1 120982 56 58 28 1 176508 54 60 38 0 179321 89 108 30 1 123185 40 49 22 1 52746 25 0 26 1 385534 92 121 25 0 33170 18 1 18 1 101645 63 20 11 0 149061 44 43 26 0 165446 33 69 25 0 237213 84 78 38 0 173326 88 86 44 0 133131 55 44 30 1 258873 60 104 40 0 180083 66 63 34 1 324799 154 158 47 1 230964 53 102 30 0 236785 119 77 31 0 135473 41 82 23 0 202925 61 115 36 1 215147 58 101 36 0 344297 75 80 30 1 153935 33 50 25 0 132943 40 83 39 0 174724 92 123 34 1 174415 100 73 31 1 225548 112 81 31 1 223632 73 105 33 0 124817 40 47 25 1 221698 45 105 33 0 210767 60 94 35 1 170266 62 44 42 0 260561 75 114 43 1 84853 31 38 30 0 294424 77 107 33 1 101011 34 30 13 1 215641 46 71 32 0 325107 99 84 36 0 7176 17 0 0 1 167542 66 59 28 1 106408 30 33 14 0 96560 76 42 17 1 265769 146 96 32 1 269651 67 106 30 1 149112 56 56 35 0 175824 107 57 20 0 152871 58 59 28 1 111665 34 39 28 0 116408 61 34 39 1 362301 119 76 34 0 78800 42 20 26 1 183167 66 91 39 0 277965 89 115 39 1 150629 44 85 33 0 168809 66 76 28 0 24188 24 8 4 0 329267 259 79 39 0 65029 17 21 18 1 101097 64 30 14 0 218946 41 76 29 0 244052 68 101 44 0 341570 168 94 21 1 103597 43 27 16 0 233328 132 92 28 1 256462 105 123 35 1 206161 71 75 28 1 311473 112 128 38 0 235800 94 105 23 0 177939 82 55 36 0 207176 70 56 32 1 196553 57 41 29 1 174184 53 72 25 1 143246 103 67 27 1 187559 121 75 36 0 187681 62 114 28 0 119016 52 118 23 0 182192 52 77 40 0 73566 32 22 23 1 194979 62 66 40 1 167488 45 69 28 1 143756 46 105 34 1 275541 63 116 33 0 243199 75 88 28 0 182999 88 73 34 1 135649 46 99 30 1 152299 53 62 33 1 120221 37 53 22 1 346485 90 118 38 1 145790 63 30 26 1 193339 78 100 35 0 80953 25 49 8 0 122774 45 24 24 0 130585 46 67 29 0 112611 41 46 20 0 286468 144 57 29 0 241066 82 75 45 1 148446 91 135 37 1 204713 71 68 33 0 182079 63 124 33 1 140344 53 33 25 1 220516 62 98 32 0 243060 63 58 29 1 162765 32 68 28 0 182613 39 81 28 0 232138 62 131 31 1 265318 117 110 52 1 85574 34 37 21 1 310839 92 130 24 1 225060 93 93 41 0 232317 54 118 33 0 144966 144 39 32 0 43287 14 13 19 1 155754 61 74 20 0 164709 109 81 31 0 201940 38 109 31 1 235454 73 151 32 0 220801 75 51 18 1 99466 50 28 23 0 92661 61 40 17 0 133328 55 56 20 0 61361 77 27 12 1 125930 75 37 17 1 100750 72 83 30 0 224549 50 54 31 0 82316 32 27 10 1 102010 53 28 13 0 101523 42 59 22 0 243511 71 133 42 1 22938 10 12 1 1 41566 35 0 9 1 152474 65 106 32 1 61857 25 23 11 0 99923 66 44 25 0 132487 41 71 36 0 317394 86 116 31 0 21054 16 4 0 1 209641 42 62 24 0 22648 19 12 13 1 31414 19 18 8 1 46698 45 14 13 0 131698 65 60 19 1 91735 35 7 18 1 244749 95 98 33 1 184510 49 64 40 1 79863 37 29 22 1 128423 64 32 38 0 97839 38 25 24 0 38214 34 16 8 0 151101 32 48 35 1 272458 65 100 43 1 172494 52 46 43 1 108043 62 45 14 1 328107 65 129 41 0 250579 83 130 38 1 351067 95 136 45 0 158015 29 59 31 1 98866 18 25 13 1 85439 33 32 28 1 229242 247 63 31 1 351619 139 95 40 1 84207 29 14 30 1 120445 118 36 16 1 324598 110 113 37 0 131069 67 47 30 1 204271 42 92 35 0 165543 65 70 32 0 141722 94 19 27 0 116048 64 50 20 1 250047 81 41 18 0 299775 95 91 31 1 195838 67 111 31 1 173260 63 41 21 1 254488 83 120 39 0 104389 45 135 41 1 136084 30 27 13 1 199476 70 87 32 0 92499 32 25 18 0 224330 83 131 39 1 135781 31 45 14 1 74408 67 29 7 1 81240 66 58 17 0 14688 10 4 0 1 181633 70 47 30 1 271856 103 109 37 1 7199 5 7 0 1 46660 20 12 5 0 17547 5 0 1 1 133368 36 37 16 0 95227 34 37 32 0 152601 48 46 24 0 98146 40 15 17 0 79619 43 42 11 1 59194 31 7 24 1 139942 42 54 22 1 118612 46 54 12 1 72880 33 14 19 0 65475 18 16 13 0 99643 55 33 17 0 71965 35 32 15 0 77272 59 21 16 1 49289 19 15 24 1 135131 66 38 15 0 108446 60 22 17 1 89746 36 28 18 1 44296 25 10 20 0 77648 47 31 16 1 181528 54 32 16 0 134019 53 32 18 1 124064 40 43 22 1 92630 40 27 8 0 121848 39 37 17 1 52915 14 20 18 1 81872 45 32 16 0 58981 36 0 23 1 53515 28 5 22 0 60812 44 26 13 1 56375 30 10 13 1 65490 22 27 16 0 80949 17 11 16 1 76302 31 29 20 0 104011 55 25 22 1 98104 54 55 17 1 67989 21 23 18 0 30989 14 5 17 1 135458 81 43 12 1 73504 35 23 7 0 63123 43 34 17 1 61254 46 36 14 1 74914 30 35 23 1 31774 23 0 17 1 81437 38 37 14 1 87186 54 28 15 0 50090 20 16 17 0 65745 53 26 21 0 56653 45 38 18 0 158399 39 23 18 1 46455 20 22 17 1 73624 24 30 17 1 38395 31 16 16 0 91899 35 18 15 1 139526 151 28 21 0 52164 52 32 16 1 51567 30 21 14 0 70551 31 23 15 0 84856 29 29 17 1 102538 57 50 15 1 86678 40 12 15 0 85709 44 21 10 1 34662 25 18 6 0 150580 77 27 22 0 99611 35 41 21 0 19349 11 13 1 1 99373 63 12 18 0 86230 44 21 17 0 30837 19 8 4 0 31706 13 26 10 0 89806 42 27 16 0 62088 38 13 16 1 40151 29 16 9 0 27634 20 2 16 0 76990 27 42 17 0 37460 20 5 7 0 54157 19 37 15 0 49862 37 17 14 1 84337 26 38 14 0 64175 42 37 18 1 59382 49 29 12 1 119308 30 32 16 1 76702 49 35 21 0 103425 67 17 19 0 70344 28 20 16 0 43410 19 7 1 0 104838 49 46 16 1 62215 27 24 10 0 69304 30 40 19 1 53117 22 3 12 0 19764 12 10 2 0 86680 31 37 14 0 84105 20 17 17 0 77945 20 28 19 1 89113 39 19 14 0 91005 29 29 11 0 40248 16 8 4 0 64187 27 10 16 0 50857 21 15 20 1 56613 19 15 12 1 62792 35 28 15 0 72535 14 17 16 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = kendall ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = kendall ; par2 = ; par3 = ; par4 = ; par5 = ; par6 = ; par7 = ; par8 = ; par9 = ; par10 = ; par11 = ; par12 = ; par13 = ; par14 = ; par15 = ; par16 = ; par17 = ; par18 = ; par19 = ; par20 = ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
panel.tau <- function(x, y, digits=2, prefix='', cex.cor) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||