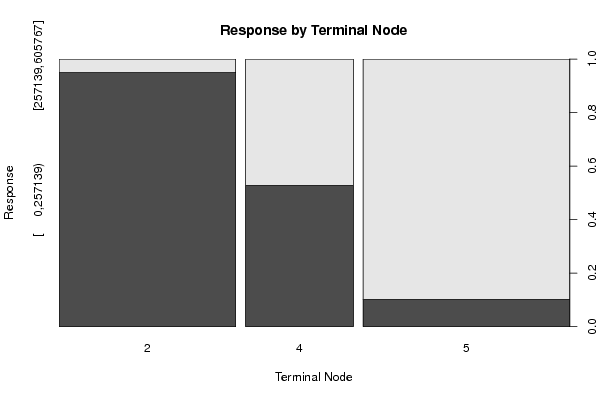Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_regression_trees1.wasp | ||||||||||||||||||||||||||||||||||||||||||||
| Title produced by software | Recursive Partitioning (Regression Trees) | ||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Fri, 23 Dec 2011 13:52:09 -0500 | ||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2011/Dec/23/t1324666343si7t3sst7v51u4g.htm/, Retrieved Mon, 29 Apr 2024 19:11:09 +0000 | ||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=160646, Retrieved Mon, 29 Apr 2024 19:11:09 +0000 | |||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 98 | ||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||
| - [Recursive Partitioning (Regression Trees)] [] [2010-12-05 20:13:50] [b98453cac15ba1066b407e146608df68] - R PD [Recursive Partitioning (Regression Trees)] [Recursive Partiti...] [2011-12-13 17:55:41] [570fce4db58fd7864ac807c4286d6e49] - R PD [Recursive Partitioning (Regression Trees)] [] [2011-12-23 13:44:42] [80bca13c5f9401fbb753952fd2952f4a] - P [Recursive Partitioning (Regression Trees)] [] [2011-12-23 18:52:09] [204816f6f70a8d342ddc2b9d4f4a80d3] [Current] | |||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||
279055 73 3 96 130 212408 75 4 75 143 233939 83 16 70 118 222117 106 2 134 146 179751 55 1 72 73 70849 28 3 8 89 605767 135 0 173 146 33186 19 0 1 22 227332 62 7 88 132 258874 48 0 98 92 359064 120 0 112 147 264989 131 7 125 203 212638 87 10 57 113 368577 85 4 139 171 269455 88 10 87 87 397992 190 0 176 208 335567 76 8 114 153 428322 172 4 121 97 182016 58 3 103 95 267365 89 8 135 197 279428 73 0 123 160 508849 111 1 99 148 206722 47 5 74 84 200004 58 9 103 227 257139 133 1 158 154 270941 138 0 116 151 324969 134 5 114 142 329962 92 0 150 148 190867 60 0 64 110 393860 79 0 150 149 327660 89 3 143 179 269239 83 6 50 149 391045 105 1 145 187 130446 49 4 56 153 430118 104 4 141 163 273950 56 0 83 127 428077 128 0 112 151 254312 93 2 79 100 120351 35 1 33 46 395643 211 2 152 156 345875 86 10 126 128 216827 82 10 97 111 224524 83 5 84 119 182485 69 6 68 148 157164 85 1 50 65 459455 157 2 101 134 78800 42 2 20 66 217932 84 0 101 201 368086 123 10 150 177 230299 70 3 129 156 244782 81 0 99 158 24188 24 0 8 7 400109 334 8 88 175 65029 17 5 21 61 101097 64 3 30 41 309810 67 1 102 133 369627 90 5 163 228 367127 204 6 132 140 377704 154 0 161 155 280106 90 12 90 141 400971 153 10 160 181 315924 122 12 139 75 291391 124 11 104 97 295075 93 8 103 142 280018 81 3 66 136 267432 71 0 163 87 217181 141 6 93 140 258166 159 10 85 169 260919 87 2 150 129 182961 73 5 143 92 256967 74 13 107 160 73566 32 6 22 67 272362 93 7 85 179 229056 62 2 101 90 229851 70 5 131 144 371391 91 4 140 144 398210 104 3 156 144 220419 111 6 81 134 231884 72 2 137 146 217714 72 0 102 121 200046 53 1 72 112 483074 131 1 161 145 146100 72 5 30 99 295224 109 2 120 96 80953 25 0 49 27 217384 63 0 121 77 179344 62 6 76 137 415550 221 1 85 151 389059 129 4 151 126 180679 106 1 165 159 299505 104 1 89 101 292260 84 3 168 144 199481 68 10 48 102 282361 78 1 149 135 329281 89 4 75 147 234577 48 5 107 155 297995 67 7 116 138 329583 89 0 173 113 416463 163 12 155 248 415683 119 13 165 116 297080 142 9 121 176 318283 70 0 156 140 224033 199 0 86 59 43287 14 4 13 64 238089 87 4 120 40 263322 160 0 117 98 299566 60 0 133 139 321797 95 0 169 135 193926 95 0 39 97 175138 105 0 125 142 354041 78 5 82 155 303273 91 1 148 115 23668 13 0 12 0 196743 79 0 146 103 61857 25 4 23 30 217543 54 0 87 130 440711 128 1 164 102 21054 16 0 4 0 252805 52 5 81 77 31961 22 0 18 9 360436 125 3 118 150 251948 77 7 76 163 187003 96 14 55 148 180842 58 3 62 94 38214 34 0 16 21 280392 56 3 98 151 358276 84 0 137 187 211775 67 0 50 171 447335 90 4 152 170 348017 99 0 163 145 441946 133 3 142 198 215177 43 0 80 152 130177 47 0 59 112 316128 363 4 94 173 466139 198 5 128 177 162279 62 16 63 153 416643 140 6 127 161 178322 86 5 60 115 292443 54 2 118 147 283913 100 1 110 124 244802 126 1 45 57 387072 125 9 96 144 246963 92 1 128 126 173260 63 3 41 78 346748 108 11 146 153 176654 59 5 147 196 268189 95 2 121 130 314070 112 1 185 159 1 0 9 0 0 14688 10 0 4 0 98 1 0 0 0 455 2 0 0 0 0 0 1 0 0 0 0 0 0 0 291650 94 2 85 94 415421 168 3 164 129 0 0 0 0 0 203 4 0 0 0 7199 5 0 7 0 46660 20 0 12 13 17547 5 0 0 4 121550 46 0 37 89 969 2 0 0 0 242774 75 2 62 71 | |||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||
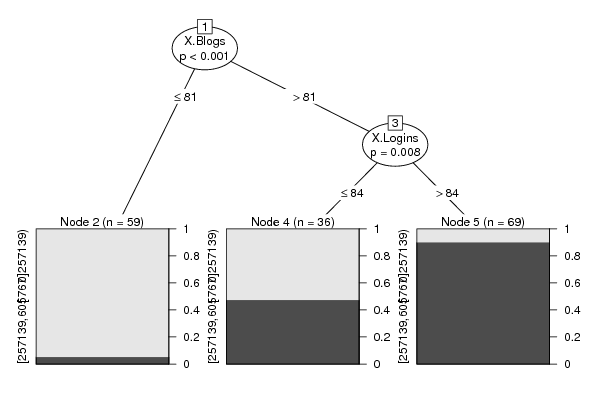
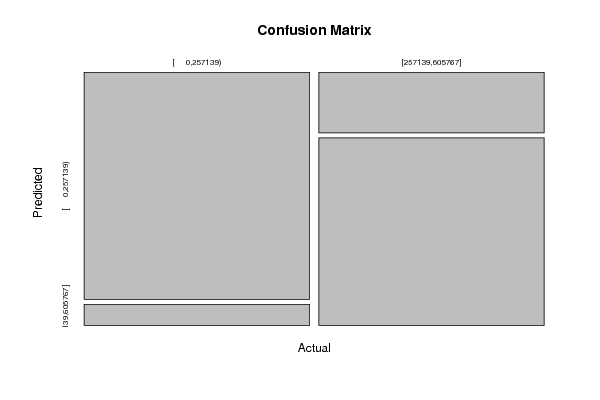
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||
| par1 = 1 ; par2 = quantiles ; par3 = 2 ; par4 = no ; | |||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||
| par1 = 1 ; par2 = quantiles ; par3 = 2 ; par4 = no ; | |||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||
library(party) | |||||||||||||||||||||||||||||||||||||||||||||