Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_regression_trees1.wasp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Title produced by software | Recursive Partitioning (Regression Trees) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Fri, 23 Dec 2011 12:15:20 -0500 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2011/Dec/23/t1324660541hec4l3ve4nfwdbs.htm/, Retrieved Mon, 29 Apr 2024 20:06:00 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=160597, Retrieved Mon, 29 Apr 2024 20:06:00 +0000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 80 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| - [One-Way-Between-Groups ANOVA- Free Statistics Software (Calculator)] [] [2010-11-02 14:17:22] [b98453cac15ba1066b407e146608df68] - RMPD [Multiple Regression] [Paper - deel 3 - ...] [2011-12-23 16:13:52] [ae1339cb5a7cf28362d01e7220b4a16c] - RMPD [Kendall tau Correlation Matrix] [Paper - Deel 3 Ke...] [2011-12-23 16:54:13] [ae1339cb5a7cf28362d01e7220b4a16c] - R D [Kendall tau Correlation Matrix] [Paper - deel3 - k...] [2011-12-23 17:00:55] [ae1339cb5a7cf28362d01e7220b4a16c] - D [Kendall tau Correlation Matrix] [Paper - deel3 pea...] [2011-12-23 17:11:23] [ae1339cb5a7cf28362d01e7220b4a16c] - RM [Recursive Partitioning (Regression Trees)] [Regression tree w...] [2011-12-23 17:15:20] [e598b5cd83fcb010b35e92a01f5e81e9] [Current] - R [Recursive Partitioning (Regression Trees)] [deel 3 regression...] [2011-12-23 18:26:42] [ae1339cb5a7cf28362d01e7220b4a16c] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
140824 95 3 96 42 130 186099 110459 68 4 75 38 143 113854 105079 64 16 70 46 118 99776 112098 139 2 134 42 146 106194 43929 51 1 83 30 73 100792 76173 46 3 8 35 89 47552 187326 118 0 173 40 146 250931 22807 46 0 1 18 22 6853 144408 79 7 88 38 132 115466 66485 76 0 104 37 92 110896 79089 82 0 114 46 147 169351 81625 66 7 125 60 203 94853 68788 60 10 57 37 113 72591 103297 117 4 139 55 171 101345 69446 50 10 87 44 87 113713 114948 133 0 176 63 208 165354 167949 63 8 114 40 153 164263 125081 100 4 121 43 97 135213 125818 44 3 103 32 95 111669 136588 65 8 135 52 197 134163 112431 103 0 123 49 160 140303 103037 103 1 99 41 148 150773 82317 62 5 77 25 84 111848 118906 70 9 103 57 227 102509 83515 159 1 158 45 154 96785 104581 78 0 116 42 151 116136 103129 101 5 114 45 142 158376 83243 73 0 150 43 148 153990 37110 58 0 64 36 110 64057 113344 147 0 150 45 149 230054 139165 54 3 143 50 179 184531 86652 84 6 50 50 149 114198 112302 56 1 145 51 187 198299 69652 45 4 56 42 153 33750 119442 87 4 141 44 163 189723 69867 87 0 83 42 127 100826 101629 77 0 112 44 151 188355 70168 72 2 79 40 100 104470 31081 36 1 33 17 46 58391 103925 51 2 152 43 156 164808 92622 44 10 126 41 128 134097 79011 75 10 97 41 111 80238 93487 87 5 84 40 119 133252 64520 97 6 68 49 148 54518 93473 90 1 50 52 65 121850 114360 860 2 101 42 134 79367 33032 57 2 20 26 66 56968 96125 99 1 107 59 201 106314 151911 120 10 150 50 177 191889 89256 76 3 129 50 156 104864 95676 56 0 99 47 158 160792 5950 20 0 8 4 7 15049 149695 94 8 88 51 175 191179 32551 21 5 21 18 61 25109 31701 70 3 30 14 41 45824 100087 133 1 102 41 133 129711 169707 86 5 166 61 228 210012 150491 224 6 132 40 140 194679 120192 65 0 161 44 155 197680 95893 86 12 90 40 141 81180 151715 70 10 160 51 181 197765 176225 148 12 139 29 75 214738 59900 72 11 104 43 97 96252 104767 59 8 103 42 142 124527 114799 67 3 66 41 136 153242 72128 58 0 163 30 87 145707 143592 60 6 93 39 140 113963 89626 105 10 85 51 169 134904 131072 84 2 154 40 129 114268 126817 63 5 143 29 92 94333 81351 67 13 107 47 160 102204 22618 39 6 22 23 67 23824 88977 60 7 85 48 179 111563 92059 94 2 101 38 90 91313 81897 67 5 131 42 144 89770 108146 96 4 140 46 144 100125 126372 54 3 156 40 144 165278 249771 54 6 81 45 134 181712 71154 62 2 137 42 146 80906 71571 71 0 102 41 121 75881 55918 50 1 74 37 112 83963 160141 117 1 161 47 145 175721 38692 45 5 30 26 99 68580 102812 61 2 120 48 96 136323 56622 31 0 49 8 27 55792 15986 175 0 121 27 77 25157 123534 70 6 76 38 137 100922 108535 284 1 85 41 151 118845 93879 95 4 151 61 126 170492 144551 72 1 165 45 159 81716 56750 63 1 89 41 101 115750 127654 75 3 168 42 144 105590 65594 90 10 48 35 102 92795 59938 89 1 149 36 135 82390 146975 138 4 75 40 147 135599 165904 68 5 107 40 155 127667 169265 80 7 116 38 138 163073 183500 65 0 181 43 113 211381 165986 130 12 155 65 248 189944 184923 85 13 165 33 116 226168 140358 83 9 121 51 176 117495 149959 89 0 176 45 140 195894 57224 116 0 86 36 59 80684 43750 43 4 13 19 64 19630 48029 87 4 120 25 40 88634 104978 80 0 117 44 98 139292 100046 132 0 133 45 139 128602 101047 59 0 169 44 135 135848 197426 50 0 39 35 97 178377 160902 87 0 125 46 142 106330 147172 62 5 82 44 155 178303 109432 70 1 148 45 115 116938 1168 9 0 12 1 0 5841 83248 54 0 146 40 103 106020 25162 25 4 23 11 30 24610 45724 113 0 87 51 130 74151 110529 63 1 164 38 102 232241 855 2 0 4 0 0 6622 101382 67 5 81 30 77 127097 14116 22 0 18 8 9 13155 89506 157 3 118 43 150 160501 135356 79 7 76 48 163 91502 116066 113 14 55 49 148 24469 144244 50 3 62 32 94 88229 8773 52 0 16 8 21 13983 102153 113 3 98 43 151 80716 117440 115 0 137 52 187 157384 104128 78 0 50 53 171 122975 134238 135 4 152 49 170 191469 134047 120 0 163 48 145 231257 279488 122 3 142 56 198 258287 79756 54 0 80 45 152 122531 66089 63 0 59 40 112 61394 102070 162 4 94 48 173 86480 146760 162 5 128 50 177 195791 154771 107 16 63 43 153 18284 165933 146 6 127 46 161 147581 64593 77 5 60 40 115 72558 92280 87 2 118 45 147 147341 67150 192 1 110 46 124 114651 128692 75 2 46 37 57 100187 124089 131 9 96 45 144 130332 125386 67 1 128 39 126 134218 37238 37 3 41 21 78 10901 140015 61 11 146 50 153 145758 150047 127 5 147 55 196 75767 154451 58 2 121 40 130 134969 156349 71 1 185 48 159 169216 0 0 9 0 0 0 0 6023 0 0 4 0 0 7953 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 84601 72 2 85 46 94 105406 68946 123 3 164 52 129 174586 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1644 0 0 7 0 0 4245 6179 7 0 12 5 13 21509 3926 3 0 0 1 4 7670 52789 106 0 37 48 89 15673 0 0 0 0 0 0 0 100350 53 2 62 34 71 75882 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
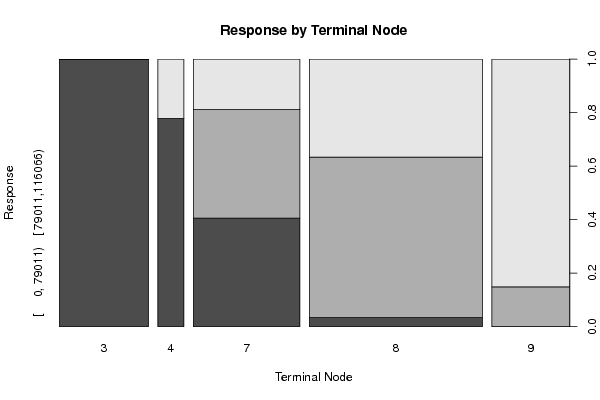
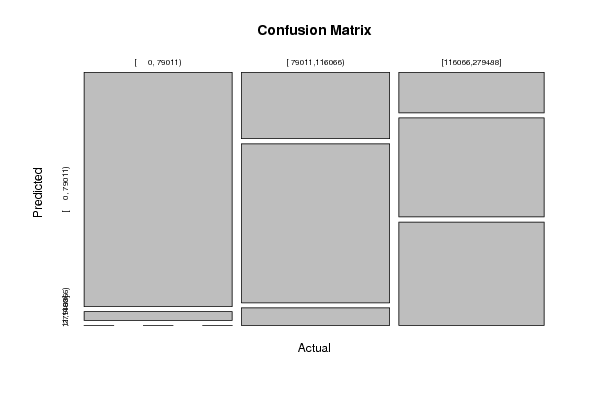
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 11 ; par2 = Do not include Seasonal Dummies ; par3 = No Linear Trend ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 1 ; par2 = quantiles ; par3 = 3 ; par4 = no ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
library(party) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||