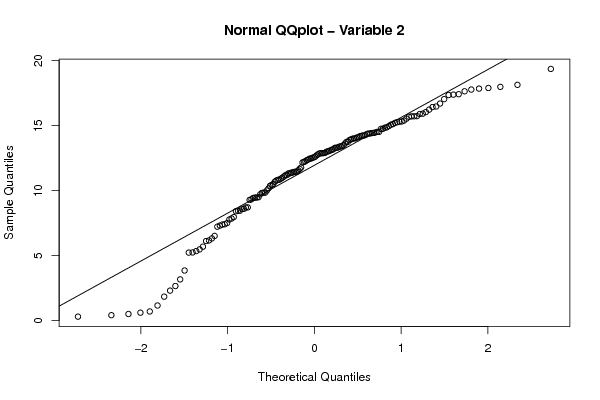
10,17463007 14,36009776
11,70685947 11,30900293
0,336629956 12,45810893
12,81545815 13,50394283
18,48134929 7,219358341
19,15 7,381367182
11,98976509 15,26096233
15,96311694 3,165415029
12,36867636 11,78143911
17,85359417 9,277808364
13,33972078 14,22780485
17,43586247 13,3753608
7,971763963 9,487750499
16,49423975 15,21511671
9,589676646 10,42712012
13,0201353 17,84978648
18,30998406 15,89534882
5,311440585 12,55115438
16,14544025 11,43396085
9,61483208 15,53507626
6,97001928 14,52239325
17,8328823 13,89331579
10,43631816 9,712278591
12,49521893 14,12942504
13,13014142 12,89537363
12,5995365 12,1602659
16,89272827 13,01181689
15,95822221 13,29873685
17,44624506 7,775415068
13,36096345 13,28429728
11,70208318 15,11451039
12,6700532 13,74797244
9,942922584 16,42942616
12,47787034 8,584377305
16,1794073 16,47410685
11,34269609 10,83539947
10,61458743 14,48035784
13,63362964 9,817153316
19,3 5,345232896
14,17862044 14,39310875
12,13110302 12,87630593
14,29122225 10,76877965
15,90424527 10,49694082
15,30537883 8,443671407
12,71711954 8,407782836
10,1041787 13,94065255
15,10633448 6,16326461
14,97472539 14,43688293
13,14940394 17,77583855
8,803370342 12,2486023
11,37671367 11,45685566
16,48829016 1,153093302
15,28690517 17,35686844
15,87449367 5,224652076
8,991891655 5,238696901
14,52069002 12,60523152
10,73329293 17,98404039
12,11255989 15,30279334
13,12039076 16,2306506
11,17618446 11,17728596
9,852452086 17,64250157
10,83826353 14,43288916
10,01602482 9,841441509
9,98429329 13,10185804
13,90654664 12,78229154
17,43829448 9,841353028
14,13651406 12,52267132
12,95465875 12,85177133
6,405875596 13,17889823
13,00827743 9,459926349
8,843896805 13,14460233
14,95275123 5,679993974
9,985399433 13,621099
15,85289071 11,4493559
6,304871788 12,45612266
11,70744131 14,52255588
14,6847188 14,00916003
9,063740747 12,87917848
14,09669644 10,91453539
11,45807414 10,67710167
14,5766967 7,500506329
8,013459929 15,71511893
10,91719297 8,451414364
9,680449615 13,04281114
12,30631181 5,475735648
19,15 6,318561991
9,974038576 11,67102852
10,61396958 12,88262502
12,39969169 12,34103121
13,89235106 15,31887313
15,3368091 10,00450285
11,27570477 14,86835659
11,46890639 8,706868119
7,705473469 12,38401963
17,80919895 14,23699191
17,4519668 12,98177075
2,080910919 14,75112225
13,09820752 15,68212424
11,29135539 18,1400865
2,507172277 15,91867058
7,823664497 14,96179352
10,96662538 16,04078615
8,284173862 7,958769343
5,69261529 6,117022453
4,687276457 7,408636579
13,25987215 12,70501195
3,580093526 12,21180573
9,733116837 13,7541755
11,88651299 10,16398398
11,3271032 11,38027871
12,99614065 14,09406222
10,51715507 15,38882422
13,71616844 0,694512385
15,32447894 10,98992814
13,82274766 3,847727223
11,71158746 10,36537965
1,908370816 15,07282766
13,82088431 0,406821823
8,241327352 11,07449882
10,5053078 2,651799226
8,180917703 13,40079885
2,322655495 14,266541
12,28598307 11,1935173
12,54906448 9,43355457
13,11109062 2,298582242
11,7934321 11,55994194
10,81286184 16,70032444
3,068772109 14,84074833
9,674855276 17,38403831
11,39973018 15,73039586
7,166265475 19,36265885
13,46103292 11,32605381
9,81357389 7,851840967
NA 13,30225262
NA 17,89762797
NA 9,46320715
NA 17,41915883
NA 9,332238773
NA 13,4187927
NA 11,37725913
NA 8,702323456
NA 14,19611665
NA 14,3980936
NA 7,301383906
NA 15,72027262
NA 14,03405468
NA 14,75498272
NA 17,03707132
NA 0,498724543
NA 10,82410397
NA 13,99003257
NA 0,295218143
NA 1,833602184
NA 0,59944272
NA 6,512322257
NA 8,594439161
|