library(MASS)
par1 <- as.numeric(par1)
par2 <- as.numeric(par2)
par2 <- round(par2,2) #rounded (we want to be able to display 10 columns)
par3 <- as.numeric(par3)
par3 <- round(par3,2) #rounded (we want to be able to display 10 columns)
par4 <- as.numeric(par4)
if (par6 == '0') par6 = 'Sturges' else par6 <- as.numeric(par6)
par7 <- as.numeric(par7)
x <- array(NA,dim=c(par7,par1))
rest.mean <- array(NA,dim=c(par7))
rest.sd <- array(NA,dim=c(par7))
rsd.mean <- array(NA,dim=c(par7))
rsd.sd <- array(NA,dim=c(par7))
for (i in 1:par7)
{
x[i,] <- rlnorm(par1,par2,par3)
x[i,] <- as.ts(x[i,]) #otherwise the fitdistr function does not work properly
dum <- fitdistr(x[i,],'log-normal')
rest.mean[i] <- dum$estimate[1]
rest.sd[i] <- dum$estimate[2]
rsd.mean[i] <- dum$sd[1]
rsd.sd[i] <- dum$sd[2]
}
nc <- par7
if (nc > 10) nc = 10
if (par5 == 'Y')
{
load(file='createtable')
a<-table.start()
a<-table.row.start(a)
a<-table.element(a,'Index',1,TRUE)
for (j in 1:nc)
{
a<-table.element(a,paste('X',j),1,TRUE)
}
a<-table.row.end(a)
if (nc < par7)
{
a<-table.row.start(a)
a<-table.element(a,'Note: only the first 10 series are displayed',nc+1,TRUE)
a<-table.row.end(a)
}
for (i in 1:par1)
{
a<-table.row.start(a)
a<-table.element(a,i,header=TRUE)
for (j in 1:nc)
{
a<-table.element(a,round(x[j,i],2))
}
a<-table.row.end(a)
}
a<-table.end(a)
table.save(a,file='mytable1.tab')
}
load(file='createtable')
a<-table.start()
a<-table.row.start(a)
a<-table.element(a,'Parameter',1,TRUE)
for (j in 1:nc)
{
a<-table.element(a,paste('X',j,sep=''),1,TRUE)
}
a<-table.row.end(a)
a<-table.row.start(a)
a<-table.element(a,' ',1,TRUE)
for (j in 1:nc)
{
a<-table.element(a,'(SD)',1,TRUE)
}
a<-table.row.end(a)
a<-table.row.start(a)
a<-table.element(a,'# simulated values',header=TRUE)
for (j in 1:nc)
{
a<-table.element(a,par1)
}
a<-table.row.end(a)
a<-table.row.start(a)
a<-table.element(a,'true mean',header=TRUE)
for (j in 1:nc)
{
a<-table.element(a,par2)
}
a<-table.row.end(a)
a<-table.row.start(a)
a<-table.element(a,'true standard deviation',header=TRUE)
for (j in 1:nc)
{
a<-table.element(a,par3)
}
a<-table.row.end(a)
a<-table.row.start(a)
a<-table.element(a,'mean',header=TRUE)
for (j in 1:nc)
{
a<-table.element(a,round(rest.mean[j],2))
}
a<-table.row.end(a)
a<-table.row.start(a)
a<-table.element(a,' ',header=TRUE)
for (j in 1:nc)
{
a<-table.element(a,round(rsd.mean[j],2))
}
a<-table.row.end(a)
a<-table.row.start(a)
a<-table.element(a,'standard deviation',header=TRUE)
for (j in 1:nc)
{
a<-table.element(a,round(rest.sd[j],2))
}
a<-table.row.end(a)
a<-table.row.start(a)
a<-table.element(a,' ',header=TRUE)
for (j in 1:nc)
{
a<-table.element(a,round(rsd.sd[j],2))
}
a<-table.row.end(a)
a<-table.end(a)
table.save(a,file='mytable.tab')
bitmap(file='test0.png')
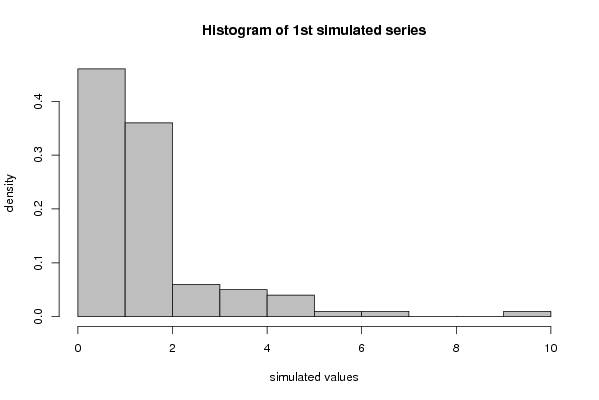
myhist<-hist(x[1,],col=par4,breaks=par6,main='Histogram of 1st simulated series',ylab='density',xlab='simulated values',freq=F)
dev.off()
bitmap(file='test1.png')
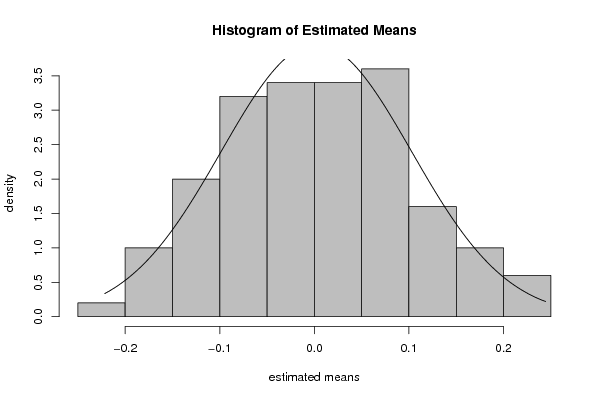
myhist<-hist(rest.mean[],col=par4,breaks=par6,main='Histogram of Estimated Means',ylab='density',xlab='estimated means',freq=F)
x <- rest.mean[]
dummean <- mean(x)
dumsd <- sd(x)
curve(1/(dumsd*sqrt(2*pi))*exp(-1/2*((x-dummean)/dumsd)^2),min(x),max(x),add=T)
dev.off()
bitmap(file='test2.png')
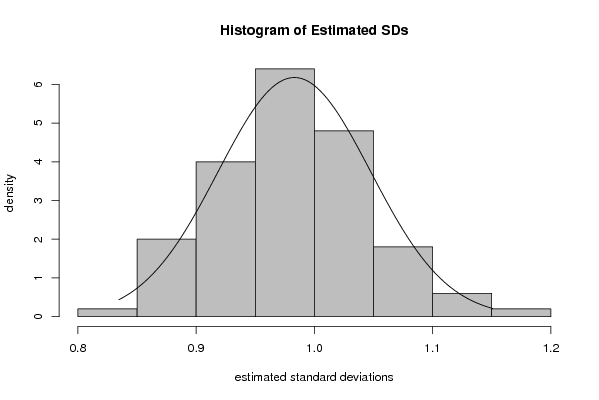
myhist<-hist(rest.sd[],col=par4,breaks=par6,main='Histogram of Estimated SDs',ylab='density',xlab='estimated standard deviations',freq=F)
x <- rest.sd[]
dummean <- mean(x)
dumsd <- sd(x)
curve(1/(dumsd*sqrt(2*pi))*exp(-1/2*((x-dummean)/dumsd)^2),min(x),max(x),add=T)
dev.off()
|