Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_linear_regression.wasp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Title produced by software | Linear Regression Graphical Model Validation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Sun, 14 Nov 2010 15:39:04 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2010/Nov/14/t1289749690u3zmpwr09x99m14.htm/, Retrieved Thu, 25 Apr 2024 19:54:58 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=94570, Retrieved Thu, 25 Apr 2024 19:54:58 +0000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 223 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| - [Linear Regression Graphical Model Validation] [Colombia Coffee -...] [2008-02-26 10:22:06] [74be16979710d4c4e7c6647856088456] - M D [Linear Regression Graphical Model Validation] [ws6vb] [2010-11-13 11:31:05] [c7506ced21a6c0dca45d37c8a93c80e0] - D [Linear Regression Graphical Model Validation] [tt] [2010-11-13 12:51:28] [4a7069087cf9e0eda253aeed7d8c30d6] - D [Linear Regression Graphical Model Validation] [W6tutorial] [2010-11-13 14:33:02] [c7506ced21a6c0dca45d37c8a93c80e0] - D [Linear Regression Graphical Model Validation] [Workshop 6, Mini ...] [2010-11-14 15:39:04] [23a9b79f355c69a75648521a893cf584] [Current] - D [Linear Regression Graphical Model Validation] [Workshop 6, Simpl...] [2010-11-16 19:47:28] [3635fb7041b1998c5a1332cf9de22bce] - R D [Linear Regression Graphical Model Validation] [Mini Toturial - H...] [2010-11-17 00:22:02] [b20a509e36241371274681d9edf773da] - R D [Linear Regression Graphical Model Validation] [] [2011-11-15 15:55:49] [46896e8a404bb9354f2d070359621409] - R D [Linear Regression Graphical Model Validation] [Mini Tutorial - k...] [2011-11-15 16:56:43] [74be16979710d4c4e7c6647856088456] - RMPD [Chi-Squared Test, McNemar Test, and Fisher Exact Test] [Chi Squared Test ...] [2011-11-15 17:42:51] [d0cddc92c01af61bef0226b9e5ade9b3] - R D [Linear Regression Graphical Model Validation] [Mini Toturial - H...] [2010-11-17 00:19:55] [b20a509e36241371274681d9edf773da] - D [Linear Regression Graphical Model Validation] [] [2011-11-15 08:53:32] [ea9976aa04c7322b215e949114660791] - R D [Linear Regression Graphical Model Validation] [] [2011-11-15 15:35:27] [46896e8a404bb9354f2d070359621409] - R D [Linear Regression Graphical Model Validation] [Mini Tutorial - t...] [2011-11-15 16:42:35] [74be16979710d4c4e7c6647856088456] - R D [Linear Regression Graphical Model Validation] [Workshop 6: Mini-...] [2011-11-15 17:30:14] [21b3d52ef28595defb5676e0f3570994] - R D [Linear Regression Graphical Model Validation] [Workshop 6: Mini-...] [2011-11-15 17:32:29] [21b3d52ef28595defb5676e0f3570994] - R D [Linear Regression Graphical Model Validation] [] [2011-12-19 16:17:39] [c505444e07acba7694d29053ca5d114e] - R D [Linear Regression Graphical Model Validation] [] [2011-12-19 16:19:11] [c505444e07acba7694d29053ca5d114e] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
5,6 4,4 2,4 4,8 3,2 4 4 4,4 6,4 4,4 5,2 4,8 3,2 4,8 4,4 1,6 3,6 3,2 3,2 5,6 6 6,4 3,6 5,6 4,4 3,2 3,6 3,6 3,6 3,6 4 6,4 4,4 3,2 3,6 6,4 4,4 6,4 4,8 4,8 5,6 3,6 4 3,6 4 4,8 5,6 5,6 4 5,6 6,4 3,6 4 2,4 3,2 5,2 4 3,2 2,8 6 3,6 4 4,8 5,2 4 4,4 3,2 3,6 5,2 4,4 3,2 3,6 3,6 6 3,6 4 5,6 4,8 4,8 4,4 5,6 2,4 4,8 3,2 5,6 4,4 4 5,6 4,8 4 5,6 2 4,4 4 3,6 4 6,4 5,2 3,6 4 4 2,8 3,6 3,2 5,6 5,6 3,2 3,6 5,6 5,6 3,2 3,2 3,2 2,8 2,4 3,2 2,4 4,4 5,6 4,4 4,4 4,4 5,6 3,2 8 4,4 3,2 4,4 4 5,6 4,4 3,6 3,6 3,2 4 5,2 5,2 4,8 3,2 5,2 5,6 4,8 5,6 6 5,2 6,4 3,6 3,6 3,6 3,2 2,8 6,4 4,4 3,6 4,4 3,6 5,6 5,2 6,4 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dataseries Y: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
5,5 3,5 8,5 5 6 6 5,5 5,5 6 6,5 7 8 5,5 5 5,5 7,5 4,5 5,5 8,5 8,5 5,5 9 7 5 5,5 7,5 7,5 6,5 8 6,5 4,5 9 9 6 8,5 4,5 4,5 6 9 6 9 7 7,5 8 5 5,5 7 4,5 6 8,5 2,5 6 6 3 12 6 6 7 3,5 6,5 6 6,5 7 4 5,5 4,5 5,5 6,5 5 5,5 6 4,5 7,5 9 7,5 6 6,5 7 5 6,5 6,5 5,5 6,5 8 4 8 5,5 4,5 8 6 7 4 4,5 7,5 5,5 10,5 7 9 6 6,5 7,5 6 9,5 7,5 5,5 5,5 5 6,5 7,5 6 6 8 4,5 9 4 6,5 8,5 4,5 7,5 4 3,5 6 7 3 4 8,5 5 5,5 7 5,5 6,5 6 5,5 4,5 6 10 6 6,5 6 6 4,5 7,5 12 3,5 8,5 5,5 8,5 5,5 6 7 5,5 8 10,5 7 10 6,5 5,5 7,5 9,5 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
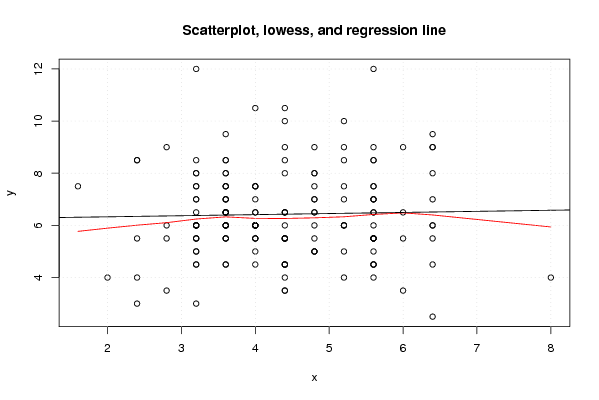
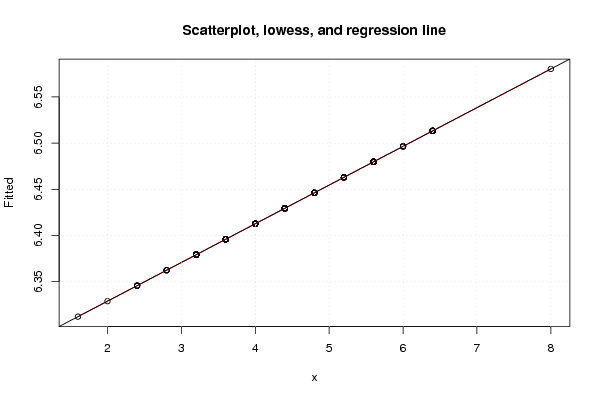
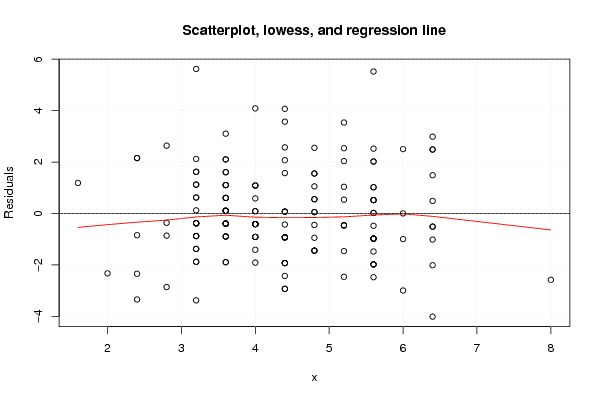
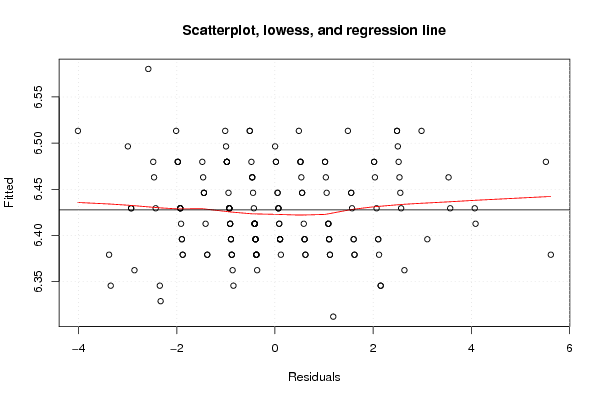
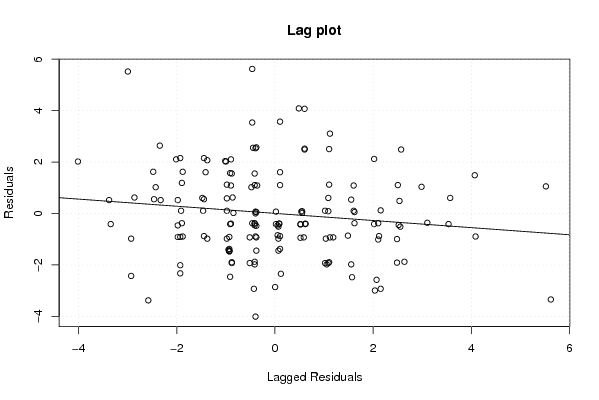
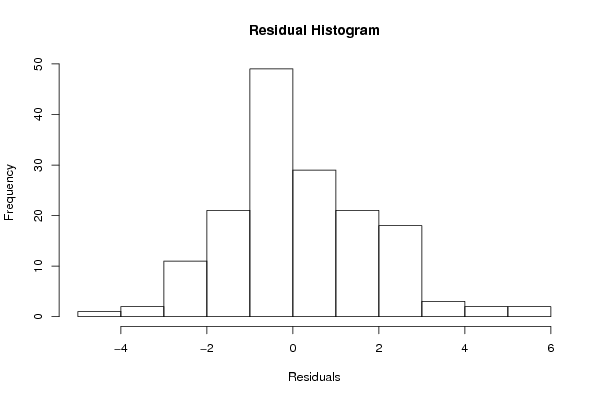
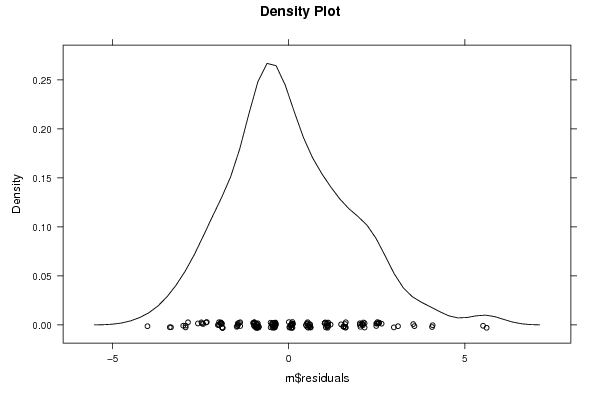
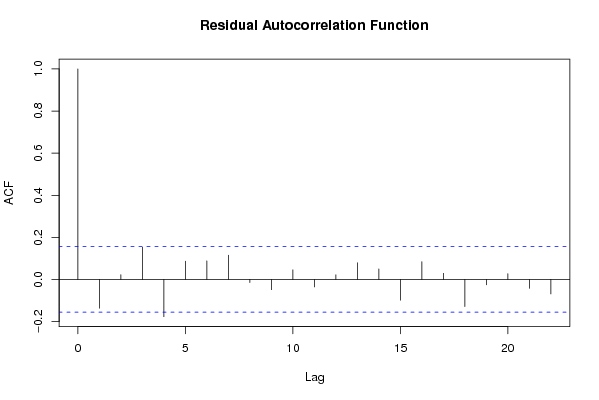
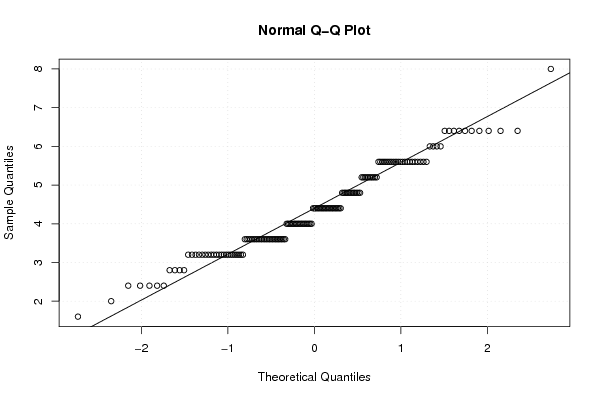
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 0 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 0 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
par1 <- as.numeric(par1) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||