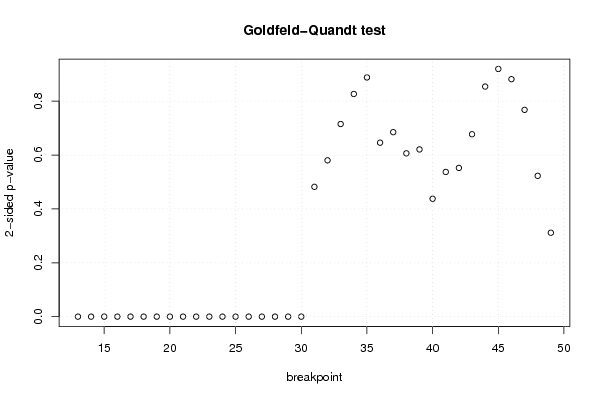
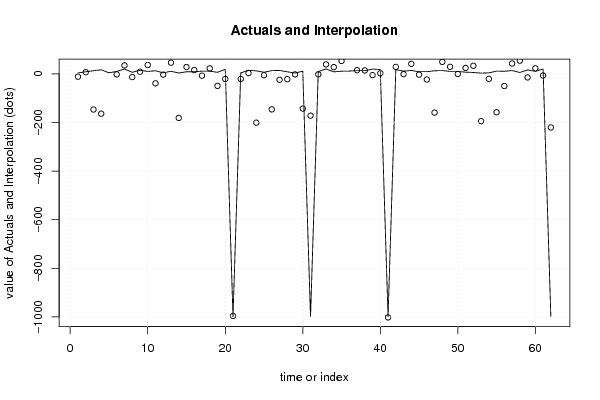
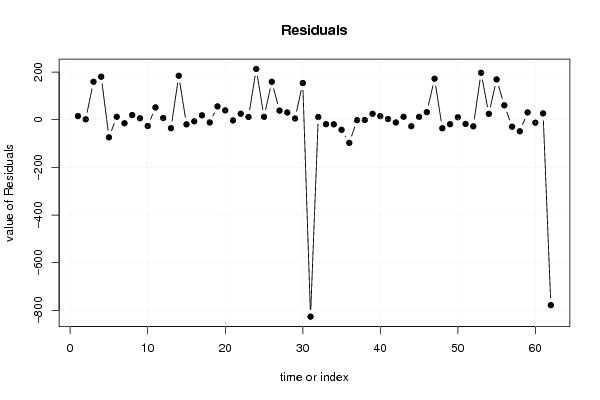
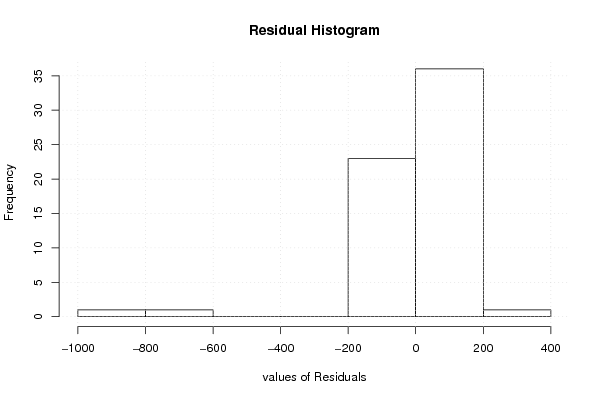
-999,00 -999,00 38,60 6654,00 5712,00 645,00 3,00 5,00 3,00 3,30
6,30 2,00 4,50 1,00 6600,00 42,00 3,00 1,00 3,00 8,30
-999,00 -999,00 14,00 3,39 44,50 60,00 1,00 1,00 1,00 12,50
-999,00 -999,00 -999,00 0,92 5,70 25,00 5,00 2,00 3,00 16,50
2,10 1,80 69,00 2547,00 4603,00 624,00 3,00 5,00 4,00 3,90
9,10 0,70 27,00 10,55 179,50 180,00 4,00 4,00 4,00 9,80
15,80 3,90 19,00 0,02 0,30 35,00 1,00 1,00 1,00 19,70
5,20 1,00 30,40 160,00 169,00 392,00 4,00 5,00 4,00 6,20
10,90 3,60 28,00 3,30 25,60 63,00 1,00 2,00 1,00 14,50
8,30 1,40 50,00 52,16 440,00 230,00 1,00 1,00 1,00 9,70
11,00 1,50 7,00 0,43 6,40 112,00 5,00 4,00 4,00 12,50
3,20 0,70 30,00 465,00 423,00 281,00 5,00 5,00 5,00 3,90
7,60 2,70 -999,00 0,55 2,40 -999,00 2,00 1,00 2,00 10,30
-999,00 -999,00 40,00 187,10 419,00 365,00 5,00 5,00 5,00 3,10
6,30 2,10 3,50 0,08 1,20 42,00 1,00 1,00 1,00 8,40
8,60 0,00 50,00 3,00 25,00 28,00 2,00 2,00 2,00 8,60
6,60 4,10 6,00 0,79 3500,00 42,00 2,00 2,00 2,00 10,70
9,50 1,20 10,40 0,20 5,00 120,00 2,00 2,00 2,00 10,70
4,80 1,30 34,00 1,41 17,50 -999,00 1,00 2,00 1,00 6,10
12,00 6,10 7,00 60,00 81,00 -999,00 1,00 1,00 1,00 18,10
-999,00 0,30 28,00 529,00 680,00 400,00 5,00 5,00 5,00 -999,00
3,30 0,50 20,00 27,66 115,00 148,00 5,00 5,00 5,00 3,80
11,00 3,40 3,90 0,12 1,00 16,00 3,00 1,00 2,00 14,40
-999,00 -999,00 39,30 207,00 406,00 252,00 1,00 4,00 1,00 12,00
4,70 1,50 41,00 85,00 325,00 310,00 1,00 3,00 1,00 6,20
-999,00 -999,00 16,20 36,33 119,50 63,00 1,00 1,00 1,00 13,00
10,40 3,40 9,00 0,10 4,00 28,00 5,00 1,00 3,00 13,80
7,40 0,80 7,60 1,04 5,50 68,00 5,00 3,00 4,00 8,20
2,10 0,80 46,00 521,00 655,00 336,00 5,00 5,00 5,00 2,90
-999,00 -999,00 22,40 100,00 157,00 100,00 1,00 1,00 1,00 10,80
-999,00 -999,00 16,30 35,00 56,00 33,00 3,00 5,00 4,00 -999,00
7,70 1,40 2,60 0,01 0,14 21,50 5,00 2,00 4,00 9,10
17,90 2,00 24,00 0,01 0,25 50,00 1,00 1,00 1,00 19,90
6,10 1,90 100,00 62,00 1320,00 267,00 1,00 1,00 1,00 8,00
8,20 2,40 -999,00 0,12 3,00 30,00 2,00 1,00 1,00 10,60
8,40 2,80 -999,00 1,35 8,10 45,00 3,00 1,00 3,00 11,20
11,90 1,30 3,20 0,02 0,40 19,00 4,00 1,00 3,00 13,20
10,80 2,00 2,00 0,05 0,33 30,00 4,00 1,00 3,00 12,80
13,80 5,60 5,00 1,70 6,30 12,00 2,00 1,00 1,00 19,40
14,30 3,10 6,50 3,50 10,80 120,00 2,00 1,00 1,00 17,40
-999,00 1,00 23,60 250,00 490,00 440,00 5,00 5,00 5,00 -999,00
15,20 1,80 12,00 0,48 15,50 140,00 2,00 2,00 2,00 17,00
10,00 0,90 20,20 10,00 115,00 170,00 4,00 4,00 4,00 10,90
11,90 1,80 13,00 1,62 11,40 17,00 2,00 1,00 2,00 13,70
6,50 1,90 27,00 192,00 180,00 115,00 4,00 4,00 4,00 8,40
7,50 0,90 18,00 2,50 12,10 31,00 5,00 5,00 5,00 8,40
-999,00 -999,00 13,70 4,29 39,20 63,00 2,00 2,00 2,00 12,50
10,60 2,60 4,70 0,28 1,90 21,00 3,00 1,00 3,00 13,20
7,40 2,40 9,80 4,24 50,40 52,00 1,00 1,00 1,00 9,80
8,40 1,20 29,00 6,80 179,00 164,00 2,00 3,00 2,00 9,60
5,70 0,90 7,00 0,75 12,30 225,00 2,00 2,00 2,00 6,60
4,90 0,50 6,00 3,60 21,00 225,00 3,00 2,00 3,00 5,40
-999,00 -999,00 17,00 14,83 98,20 150,00 5,00 5,00 5,00 2,60
3,20 0,60 20,00 55,50 175,00 151,00 5,00 5,00 5,00 3,80
-999,00 -999,00 12,70 1,40 12,50 90,00 2,00 2,00 2,00 11,00
8,10 2,20 3,50 0,06 1,00 -999,00 3,00 1,00 2,00 10,30
11,00 2,30 4,50 0,90 2,60 60,00 2,00 1,00 2,00 13,30
4,90 0,50 7,50 2,00 12,30 200,00 3,00 1,00 3,00 5,40
13,20 2,60 2,30 0,10 2,50 46,00 3,00 2,00 2,00 15,80
9,70 0,60 24,00 4,19 58,00 210,00 4,00 3,00 4,00 10,30
12,80 6,60 3,00 3,50 3,90 14,00 2,00 1,00 1,00 19,40
-999,00 -999,00 13,00 4,05 17,00 38,00 3,00 1,00 1,00 -999,00
|