Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_edauni.wasp | ||||||||||||||||||||||||||||||||||||||||||||||||||||
| Title produced by software | Univariate Explorative Data Analysis | ||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Tue, 20 Oct 2009 15:51:28 -0600 | ||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2009/Oct/20/t12560755324onat1a8ec5qyx9.htm/, Retrieved Thu, 02 May 2024 21:00:38 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=49211, Retrieved Thu, 02 May 2024 21:00:38 +0000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 132 | ||||||||||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| - [Bivariate Data Series] [Bivariate dataset] [2008-01-05 23:51:08] [74be16979710d4c4e7c6647856088456] F RMPD [Univariate Explorative Data Analysis] [Colombia Coffee] [2008-01-07 14:21:11] [74be16979710d4c4e7c6647856088456] F RMPD [Univariate Data Series] [] [2009-10-14 08:30:28] [74be16979710d4c4e7c6647856088456] - RMPD [Univariate Explorative Data Analysis] [Workshop3 - histo...] [2009-10-20 19:05:41] [df6326eec97a6ca984a853b142930499] - PD [Univariate Explorative Data Analysis] [workshop 3 vraag ...] [2009-10-20 21:51:28] [0cc924834281808eda7297686c82928f] [Current] | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||||||||||
0.355911391 0.192295904 0.272267288 0.258689189 0.18567135 0.373877063 0.34691589 0.359880227 0.286165509 0.414310922 0.482794147 0.317571037 0.346366101 -0.13487635 -0.187377539 0.285965011 -0.081923433 0.165051707 -0.012919634 0.139960994 0.073744737 0.121091076 0.028232563 0.206453634 0.257216569 0.065350838 0.05924699 -0.052759239 0.047943153 0.029757832 0.10780113 0.084875467 -0.067866223 -0.050792543 -0.160144441 -0.169203507 0.148743398 -0.082307667 -0.127673304 -0.01080757 -0.315287757 0.042567136 0.005232124 -0.170879984 -0.044496029 -0.112199933 -0.150843154 -0.252805389 0.004218806 -0.105782565 -0.155260875 -0.063622103 -0.171783206 -0.045750814 -0.108936836 -0.101035287 -0.264863038 -0.106527791 -0.128082303 -0.208906985 -0.15472301 -0.107099467 -0.290272108 0.076359113 -0.233991104 -0.127904111 -0.137345117 -0.146830587 -0.153801771 -0.039895855 -0.332238711 -0.158714763 0.002967834 -0.001027051 -0.010295419 0.034237962 -0.159297351 -0.158707613 0.110832302 -0.003154811 -0.021356073 0.112124864 0.044872239 0.017477842 0.10245901 0.098946073 -0.206535883 -0.006024052 -0.219470394 0.057492629 0.132658623 0.014386318 0.277202467 -0.051526973 0.058962934 -0.099306341 0.197363426 -0.1163449 0.136960891 -0.084933941 -0.005866992 -0.031379518 0.098016068 0.009935834 0.049865297 0.194801823 0.067147354 -0.078575054 0.0080191 0.001027051 -0.015792757 0.025921638 -0.041486393 0.07401796 0.059698617 -0.019134923 -0.008463373 0.14716635 0.122507323 0.052540291 | |||||||||||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||
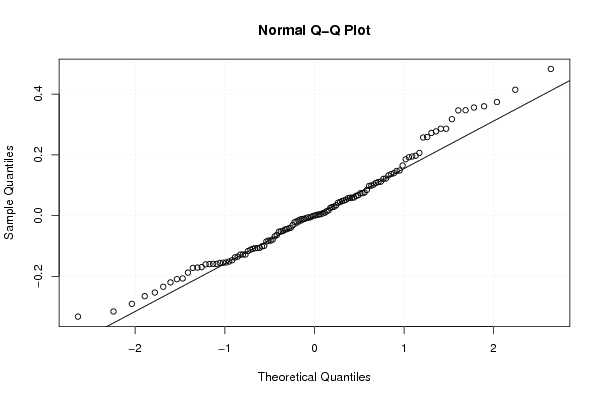
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 500 ; par2 = 12 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 0 ; par2 = 36 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||||||||||
par1 <- as.numeric(par1) | |||||||||||||||||||||||||||||||||||||||||||||||||||||