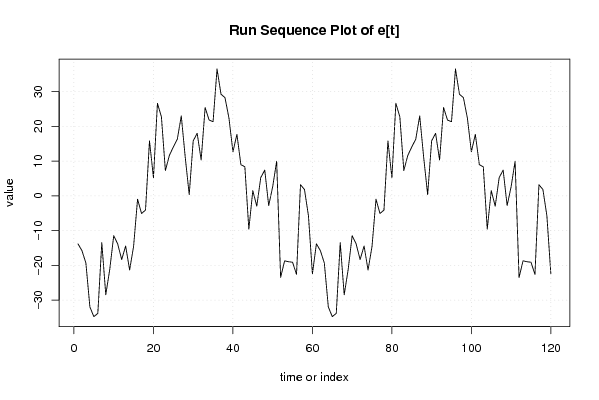
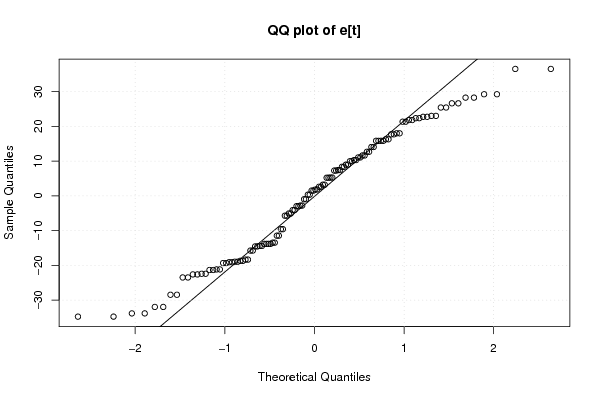
-355,051
-353,784
-354,293
-350,691
-350,155
-350,167
-349,02
-341,187
-348,266
-351,553
-349,377
-344,289
-343,656
-339,999
-344,7
-347,988
-347,58
-346,825
-345,237
-337,099
-350,251
-349,859
-345,32
-343,773
-341,272
-342,001
-346,253
-342,508
-337,708
-344,583
-338,033
-329,933
-338,326
-332,973
-335,128
-338,169
-334,62
-333,972
-334,277
-331,626
-333,494
-330,872
-322,131
-313,857
-322,424
-321,669
-328,637
-326,1
-320,355
-323,196
-330,737
-315,004
-318,118
-318,776
-310,722
-308,044
-323,375
-330,393
-331,46
-322,721
-355,051
-353,784
-354,293
-350,691
-350,155
-350,167
-349,02
-341,187
-348,266
-351,553
-349,377
-344,289
-343,656
-339,999
-344,7
-347,988
-347,58
-346,825
-345,237
-337,099
-350,251
-349,859
-345,32
-343,773
-341,272
-342,001
-346,253
-342,508
-337,708
-344,583
-338,033
-329,933
-338,326
-332,973
-335,128
-338,169
-334,62
-333,972
-334,277
-331,626
-333,494
-330,872
-322,131
-313,857
-322,424
-321,669
-328,637
-326,1
-320,355
-323,196
-330,737
-315,004
-318,118
-318,776
-310,722
-308,044
-323,375
-330,393
-331,46
-322,721 |
-7,941
-6,714
-11,573
-15,211
-16,665
-15,757
7,47
12,053
1,654
3,157
6,243
14,421
19,924
22,151
17,2
22,562
19,47
22,335
46,243
55,961
44,519
41,611
37,47
45,697
54,378
54,789
50,887
48,292
49,562
47,887
66,427
78,967
73,114
82,887
77,022
84,611
86,2
86,838
80,163
77,114
77,476
75,298
96,519
99,243
88,936
86,341
77,163
85,66
89,795
88,114
76,623
82,476
79,482
77,574
97,568
100,746
88,255
69,347
59,12
64,249
-7,941
-6,714
-11,573
-15,211
-16,665
-15,757
7,47
12,053
1,654
3,157
6,243
14,421
19,924
22,151
17,2
22,562
19,47
22,335
46,243
55,961
44,519
41,611
37,47
45,697
54,378
54,789
50,887
48,292
49,562
47,887
66,427
78,967
73,114
82,887
77,022
84,611
86,2
86,838
80,163
77,114
77,476
75,298
96,519
99,243
88,936
86,341
77,163
85,66
89,795
88,114
76,623
82,476
79,482
77,574
97,568
100,746
88,255
69,347
59,12
64,249 |