Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_edabi.wasp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Title produced by software | Bivariate Explorative Data Analysis | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Mon, 02 Nov 2009 07:58:53 -0700 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2009/Nov/02/t1257174005jzrvoge2iozk4td.htm/, Retrieved Sat, 04 May 2024 03:40:43 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=52695, Retrieved Sat, 04 May 2024 03:40:43 +0000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 200 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| - [Bivariate Data Series] [Bivariate dataset] [2008-01-05 23:51:08] [74be16979710d4c4e7c6647856088456] - PD [Bivariate Data Series] [Reproduction Part 1] [2009-10-26 18:51:43] [96e597a9107bfe8c07649cce3d4f6fec] - RMPD [Bivariate Explorative Data Analysis] [JJ Workshop 4, De...] [2009-10-26 19:42:48] [96e597a9107bfe8c07649cce3d4f6fec] - D [Bivariate Explorative Data Analysis] [JJ Workshop 4, de...] [2009-10-27 18:56:34] [96e597a9107bfe8c07649cce3d4f6fec] - RM D [Bivariate Explorative Data Analysis] [JJ Workshop 5, mo...] [2009-11-02 13:36:21] [96e597a9107bfe8c07649cce3d4f6fec] - D [Bivariate Explorative Data Analysis] [JJ Workshop 5, mo...] [2009-11-02 14:58:53] [e31f2fa83f4a5291b9a51009566cf69b] [Current] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-11.152 -1.5796 13.16 11.1072 -5.6192 -3.8776 13.7234 7.109 10.9492 7.6378 2.6788 2.4724 -10.7344 0.574 15.1298 4.6936 -11.3734 -18.5268 1.4216 -4.6246 -4.2308 8.5854 -2.922 0.5006 -9.2324 0.2036 5.65 4.1602 -15.015 -20.315 2.9446 -6.449 -7.137 3.139 -9.8002 -4.357 -6.117 0.6406 0.589 2.3694 -15.0224 -19.9452 6.3754 -5.8596 -3.3378 -1.0326 -4.4502 -4.275 -3.768 11.3846 18.4804 18.5824 -4.7104 -2.5836 5.172 4.139 9.136 15.4814 1.9896 2.7668 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dataseries Y: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-1911,724 -1092,9502 2107,62 1781,9164 -2191,0104 -3196,1112 1042,9483 -510,7845 1350,1454 -310,0889 528,2906 213,2038 -1599,2628 1127,863 3179,2851 938,3232 -867,7033 -3318,9066 1899,2492 -28,0477 -706,9446 2361,3973 -531,939 -326,1803 -1150,7638 1250,9782 2070,885 1756,2899 -1364,0325 -4171,1225 1808,3077 598,8945 -1071,6315 2335,7905 -846,1699 -1114,4615 -301,8415 1165,2397 581,1755 2189,5053 -1432,1288 -3959,7774 2481,9723 -904,7102 -68,7611 292,8563 -1419,0249 -1625,1425 -95,996 360,3377 1786,0998 2683,3188 -953,3848 -2681,8982 2139,734 -776,0495 258,852 2343,5493 -914,6248 -1193,5834 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
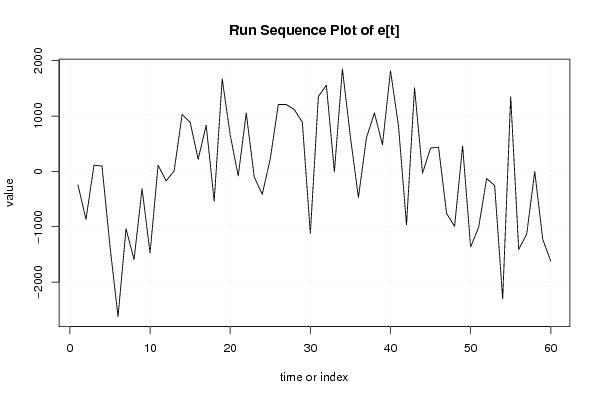
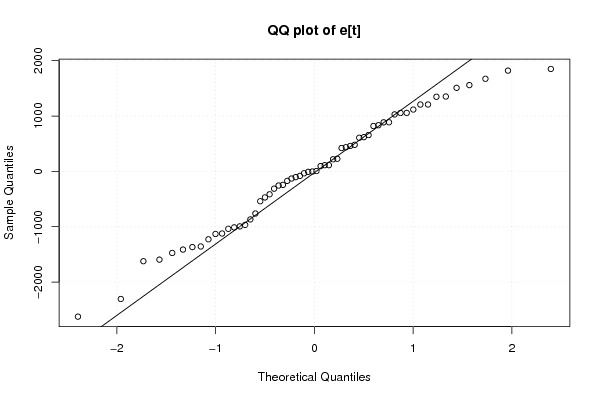
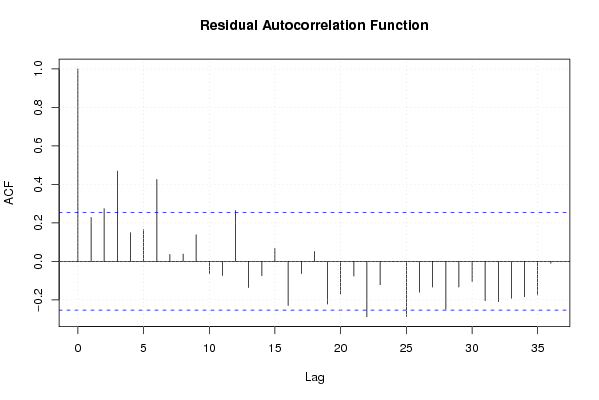
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 0 ; par2 = 36 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 0 ; par2 = 36 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
par1 <- as.numeric(par1) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||