Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_smp.wasp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
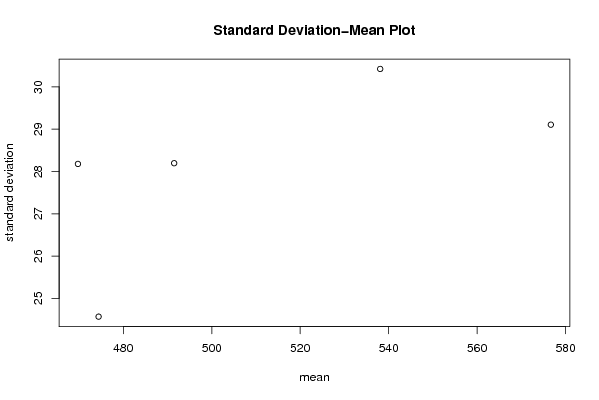
| Title produced by software | Standard Deviation-Mean Plot | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Fri, 05 Dec 2008 03:14:55 -0700 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2008/Dec/05/t1228472718a5pn1ys93gmm5rz.htm/, Retrieved Thu, 16 May 2024 17:14:21 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=29137, Retrieved Thu, 16 May 2024 17:14:21 +0000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 263 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| - [Standard Deviation-Mean Plot] [Paper Hoofdstuk 4...] [2008-12-05 10:14:55] [286e96bd53289970f8e5f25a93fb50b3] [Current] - RM [Variance Reduction Matrix] [Paper Hoofdstuk 4...] [2008-12-05 10:50:46] [6fea0e9a9b3b29a63badf2c274e82506] - RMP [(Partial) Autocorrelation Function] [Paper Hoofdstuk 4...] [2008-12-05 11:00:50] [6fea0e9a9b3b29a63badf2c274e82506] - RMP [(Partial) Autocorrelation Function] [Paper Hoofdstuk 4...] [2008-12-05 11:12:26] [6fea0e9a9b3b29a63badf2c274e82506] - RMP [(Partial) Autocorrelation Function] [Paper Hoofdstuk 4...] [2008-12-05 11:12:26] [6fea0e9a9b3b29a63badf2c274e82506] - RMP [(Partial) Autocorrelation Function] [Paper Hoofdstuk 4...] [2008-12-05 11:21:04] [6fea0e9a9b3b29a63badf2c274e82506] - P [(Partial) Autocorrelation Function] [Paper Hoofdstuk 4...] [2008-12-05 22:37:51] [6fea0e9a9b3b29a63badf2c274e82506] - P [(Partial) Autocorrelation Function] [Paper Hoofdstuk 4...] [2008-12-05 23:07:43] [6fea0e9a9b3b29a63badf2c274e82506] - RMP [Spectral Analysis] [Paper Hoofdstuk 4...] [2008-12-05 23:23:55] [6fea0e9a9b3b29a63badf2c274e82506] - P [Spectral Analysis] [Paper Hoofdstuk 4...] [2008-12-06 11:37:54] [6fea0e9a9b3b29a63badf2c274e82506] - P [Spectral Analysis] [Paper Hoofdstuk 4...] [2008-12-06 11:44:44] [6fea0e9a9b3b29a63badf2c274e82506] - D [Standard Deviation-Mean Plot] [Paper, hoofdstuk ...] [2008-12-06 15:40:41] [79c17183721a40a589db5f9f561947d8] - RM D [Variance Reduction Matrix] [Paper, hoofdstuk ...] [2008-12-06 15:48:43] [79c17183721a40a589db5f9f561947d8] - RMP [(Partial) Autocorrelation Function] [Paper, hoofdstuk ...] [2008-12-06 16:03:27] [79c17183721a40a589db5f9f561947d8] - RMP [(Partial) Autocorrelation Function] [Paper, hoofdstuk ...] [2008-12-06 16:56:09] [79c17183721a40a589db5f9f561947d8] - P [(Partial) Autocorrelation Function] [Paper, hoofdstuk ...] [2008-12-06 17:05:19] [79c17183721a40a589db5f9f561947d8] - RMP [Spectral Analysis] [Paper, hoofdstuk ...] [2008-12-07 11:35:08] [79c17183721a40a589db5f9f561947d8] - P [Spectral Analysis] [Paper, hoofdstuk ...] [2008-12-07 11:55:02] [79c17183721a40a589db5f9f561947d8] - P [Spectral Analysis] [Paper, hoofdstuk ...] [2008-12-07 12:00:47] [79c17183721a40a589db5f9f561947d8] - P [Spectral Analysis] [Paper 4.4 Spectru...] [2008-12-16 14:34:09] [79c17183721a40a589db5f9f561947d8] - P [Spectral Analysis] [Paper 4.4 Spectru...] [2008-12-16 14:37:47] [79c17183721a40a589db5f9f561947d8] - P [Spectral Analysis] [Paper 4.4 Spectru...] [2008-12-16 14:39:52] [79c17183721a40a589db5f9f561947d8] - RMP [(Partial) Autocorrelation Function] [Paper, hoofdstuk ...] [2008-12-07 12:59:04] [79c17183721a40a589db5f9f561947d8] - P [(Partial) Autocorrelation Function] [Paper, hoofdstuk ...] [2008-12-09 14:22:25] [79c17183721a40a589db5f9f561947d8] - RMPD [ARIMA Backward Selection] [Paper Hoofdstuk 4...] [2008-12-09 14:31:45] [6fea0e9a9b3b29a63badf2c274e82506] - RMP [ARIMA Backward Selection] [Paper Hoofdstuk 4...] [2008-12-09 14:42:05] [79c17183721a40a589db5f9f561947d8] - [ARIMA Backward Selection] [Paper, hoofdstuk ...] [2008-12-10 20:45:33] [79c17183721a40a589db5f9f561947d8] - P [(Partial) Autocorrelation Function] [Paper 4.3 (P)ACF ...] [2008-12-16 14:21:32] [79c17183721a40a589db5f9f561947d8] - P [(Partial) Autocorrelation Function] [Paper 4.3 (P)ACF ...] [2008-12-16 14:24:05] [79c17183721a40a589db5f9f561947d8] - [(Partial) Autocorrelation Function] [Paper 4.3 (P)ACF ...] [2008-12-16 14:28:29] [79c17183721a40a589db5f9f561947d8] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
493.000 481.000 462.000 457.000 442.000 439.000 488.000 521.000 501.000 485.000 464.000 460.000 467.000 460.000 448.000 443.000 436.000 431.000 484.000 510.000 513.000 503.000 471.000 471.000 476.000 475.000 470.000 461.000 455.000 456.000 517.000 525.000 523.000 519.000 509.000 512.000 519.000 517.000 510.000 509.000 501.000 507.000 569.000 580.000 578.000 565.000 547.000 555.000 562.000 561.000 555.000 544.000 537.000 543.000 594.000 611.000 613.000 611.000 594.000 595.000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 12 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 12 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
par1 <- as.numeric(par1) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||